deseq_analysis.r 差异基因分析DESeq2
deseq_analysis.r 差异基因分析DESeq2
使用方法:
$Rscript $scriptdir/deseq_analysis.r -h
usage: /work/my_stad_immu/scripts/deseq_analysis.r [-h] -i filepath -m
filepath -t treatname
--control CONTROL --case
CASE [-f fdr] [-c fc]
[-s size] [-a alpha]
[-X x.lab] [-Y y.lab]
[-T title] [-H height]
[-W width] [-o path]
[-p prefix]
DESeq2 analysis : https://www.omicsclass.com/article/1500
optional arguments:
-h, --help show this help message and exit
-i filepath, --input filepath
input read count file [required]
-m filepath, --metadata filepath
metadata file , required
-t treatname, --treatname treatname
treat colname in group file, required
--control CONTROL set control group name required
--case CASE set case group name required
-f fdr, --fdr fdr set fdr threshold [default 0.05]
-c fc, --fc fc set fold change threshold [default 2]
-s size, --size size point size [optional, default: 0.7]
-a alpha, --alpha alpha
point transparency [0-1] [optional, default: 1]
-X x.lab, --x.lab x.lab
the label for x axis [optional, default: log2FC]
-Y y.lab, --y.lab y.lab
the label for y axis [optional, default: -log10(FDR)]
-T title, --title title
the label for main title [optional, default: Volcano]
-H height, --height height
the height of pic inches [default 5]
-W width, --width width
the width of pic inches [default 5]
-o path, --outdir path
output file directory [default
/work/my_stad_immu/05.enrich]
-p prefix, --prefix prefix
out file name prefix [default Volcano]
参数说明:
-i 输入基因表达矩阵文件,必须为count表达文件:
| ID | TCGA-B7-A5TK-01A-12R-A36D-31 | TCGA-BR-7959-01A-11R-2343-13 | TCGA-IN-8462-01A-11R-2343-13 | TCGA-BR-A4CR-01A-11R-A24K-31 | TCGA-CG-4443-01A-01R-1157-13 | TCGA-KB-A93J-01A-11R-A39E-31 | TCGA-BR-4371-01A-01R-1157-13 |
| TSPAN6 | 5951 | 4036 | 2834 | 3484 | 2537 | 2027 | 4749 |
| TNMD | 3 | 4 | 0 | 1 | 0 | 1 | 8 |
| DPM1 | 4672 | 4330 | 1725 | 4370 | 6523 | 3094 | 4415 |
| SCYL3 | 1260 | 2057 | 702 | 1483 | 924 | 1451 | 982 |
| C1orf112 | 523 | 992 | 172 | 1400 | 234 | 733 | 958 |
| FGR | 1249 | 1127 | 285 | 148 | 56 | 941 | 208 |
| CFH | 12831 | 11435 | 5387 | 995 | 4571 | 2189 | 1795 |
| FUCA2 | 5896 | 7857 | 3208 | 5625 | 1527 | 7530 | 3290 |
| GCLC | 2682 | 5509 | 1447 | 9323 | 6422 | 5265 | 2418 |
-k 输入基因表达矩阵文件,为fpkm 或者 tpm文件 用于相关性分析:
| ID | TCGA-B7-A5TK-01A-12R-A36D-31 | TCGA-BR-7959-01A-11R-2343-13 | TCGA-IN-8462-01A-11R-2343-13 | TCGA-BR-A4CR-01A-11R-A24K-31 | TCGA-CG-4443-01A-01R-1157-13 |
| TSPAN6 | 59.65411 | 32.83064 | 40.7596 | 39.84131 | 53.37611 |
| TNMD | 0.084708 | 0.091652 | 0 | 0.032211 | 0 |
| DPM1 | 175.9638 | 132.3385 | 93.21574 | 187.7617 | 515.6368 |
| SCYL3 | 8.321862 | 11.02458 | 6.652222 | 11.17368 | 12.80849 |
| C1orf112 | 3.984496 | 6.132832 | 1.880095 | 12.1676 | 3.741654 |
| FGR | 16.34408 | 11.96739 | 5.350846 | 2.209351 | 1.53802 |
-m metadata文件路径,样本的分组信息,第一列必须和表达文件的样本名称对应:
| barcode | subtype.hclust | StromalScore | ImmuneScore | ESTIMATEScore | TumourPurity |
| TCGA-B7-A5TK-01A-12R-A36D-31 | S1 | 1026.057 | 2386.835 | 3412.892 | 0.448276 |
| TCGA-BR-7959-01A-11R-2343-13 | S2 | 1130.722 | 729.402 | 1860.124 | 0.638667 |
| TCGA-IN-8462-01A-11R-2343-13 | S2 | 112.2318 | 683.9349 | 796.1667 | 0.750581 |
| TCGA-BR-A4CR-01A-11R-A24K-31 | S2 | -1060.35 | -766.618 | -1826.97 | 0.943814 |
| TCGA-CG-4443-01A-01R-1157-13 | S2 | -261.577 | -258.629 | -520.206 | 0.8635 |
| TCGA-KB-A93J-01A-11R-A39E-31 | S1 | -202.255 | 1605.12 | 1402.865 | 0.688838 |
| TCGA-BR-4371-01A-01R-1157-13 | S2 | -828.231 | 711.3379 | -116.893 | 0.832147 |
| TCGA-IN-A6RO-01A-12R-A33Y-31 | S2 | -1406.57 | 68.58307 | -1337.98 | 0.917683 |
| TCGA-HU-A4H3-01A-21R-A251-31 | S2 | -619.208 | 538.7225 | -80.4854 | 0.829171 |
| TCGA-RD-A8MV-01A-11R-A36D-31 | S1 | 113.4127 | 2309.647 | 2423.06 | 0.572976 |
| TCGA-VQ-A91X-01A-12R-A414-31 | S2 | -1845.85 | -590.017 | -2435.87 | 0.969545 |
| TCGA-D7-8575-01A-11R-2343-13 | S2 | -206.112 | 1392.799 | 1186.687 | 0.711491 |
| TCGA-BR-4257-01A-01R-1131-13 | S1 | 861.029 | 1676.148 | 2537.177 | 0.559167 |
| TCGA-BR-8485-01A-11R-2402-13 | S1 | 373.0961 | 1110.516 | 1483.612 | 0.680198 |
| TCGA-BR-4370-01A-01R-1157-13 | S1 | 1300.495 | 1802.327 | 3102.822 | 0.488483 |
-t subtype.hclust --case S1 --control S2 : 指定metadata 分组列名,分组里面的比较组名字 ,如果分组名字有空格,应该用引号引起来: “Stage IA”
--fdr 0.01 --fc 2 设置差异基因的筛选条件: 显著性和差异倍数
使用举例:
Rscript $scriptdir/deseq_analysis.r -i ../01.TCGA_download/TCGA-STAD_gene_expression_Counts.tsv \ -k ../01.TCGA_download/TCGA-STAD_gene_expression_TPM.tsv --fdr 0.01 --fc 2 \ -m ../03.TIME/metadata.group.tsv -t subtype.hclust --case S1 --control S2 -p S1_vs_S2
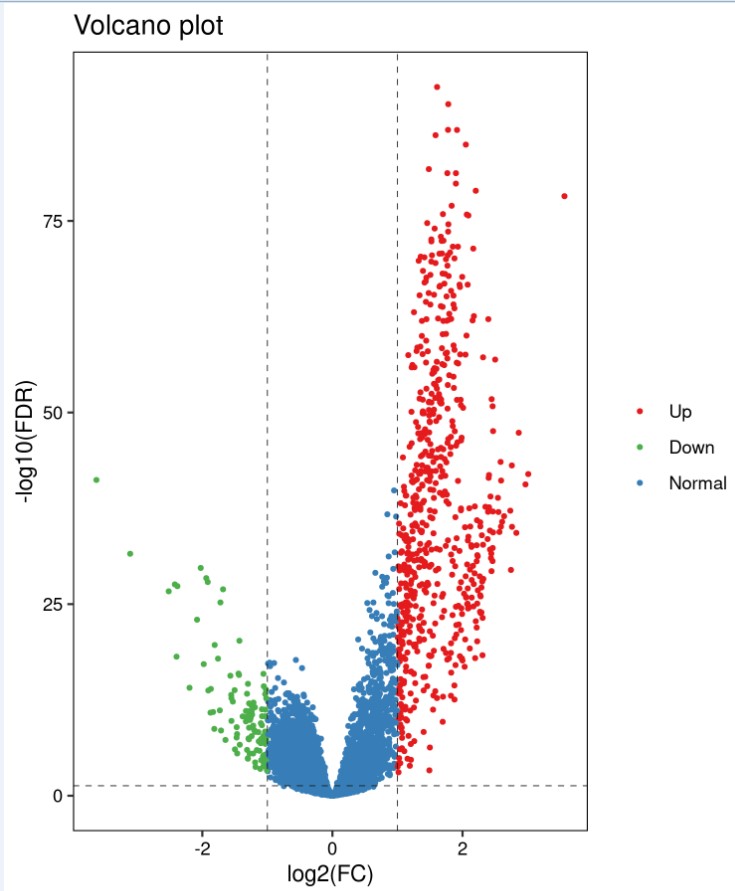
结果展示:
火山图:
参考文献:
Love MI, Huber W, Anders S (2014). “Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2.” Genome Biology, 15, 550. doi: 10.1186/s13059-014-0550-8.
- 发表于 2021-06-23 11:31
- 阅读 ( 1953 )
- 分类:转录组
你可能感兴趣的文章
相关问题
- 差异基因表达分析 1 回答
- 差异基因分析多个样本 老师我想请问一下这个代码是什么意思 分组的mata文件是什么样的 我的样品名应该怎么输入 1 回答
- 用deseq_analysis.r分析差异表达分析时报如下的错误 1 回答
- 老师,我在转录组分析的时候,遇到这个问题,怎么解决呀 1 回答
- 老师,请帮忙,感谢。结果有参转录组5.deg报错。 2 回答
- 各位大佬好!我在筛选一批转录组数据里的差异基因时,当进行到构建dds对象的时候,r语言出现以下报错,说“一些值不是整数”,当我上网查这个错误的时候,有人说是“数据已进行归一化,无法进行DESeq2分析”,请问各位大佬,还有什么解决方法嘛??谢谢谢谢 2 回答
0 条评论
请先 登录 后评论
