compare_stat_boxplot.r metadata中变量分组T检验比较并绘制box图
compare_stat_boxplot.r metadata中变量分组T检验比较并绘制box图
使用说明:
metadata中按列分组,做差异比较分析,可以利用 merge_tsv_files.r 或者 merge_metadata_genexpdata.r 将比较的信息合并到metadata中:
usage: /share/nas1/huangls/test/TCGA_immu/scripts/compare_stat_boxplot.r
[-h] -m filepath -v variate [variate ...] -g group [-b groupby]
[-G geom] [--addDot] [-p palette [palette ...]] [-T title] [-x xlab]
[-y ylab] [-o path] [-n prefix] [-H number] [-W number]
box plot and stat :
optional arguments:
-h, --help show this help message and exit
-m filepath, --metadata filepath
input metadata file path[required]
-v variate [variate ...], --variate variate [variate ...]
variate for box [required]
-g group, --group group
group name from metadata to test[required]
-b groupby, --groupby groupby
main group name from metadata to subset [default NULL]
-G geom, --geom geom set type of plot: boxplot, violin, splitviolin
[default=boxplot]
-p palette [palette ...], --palette palette [palette ...]
fill color palette [default ('#e41a1c', '#377eb8',
'#4daf4a', '#984ea3', '#ff7f00', '#ffff33', '#a65628',
'#f781bf', '#999999')]
-T title, --title title
the label for main title [optional, default: ]
-x xlab, --xlab xlab input xlab [default ]
-y ylab, --ylab ylab input ylab [default value]
-o path, --outdir path
output file directory [default
/share/nas1/huangls/test/TCGA_immu/07.survival]
-n prefix, --name prefix
out file name prefix [default demo]
-H number, --height number
the height of pic inches [default 5]
-W number, --width number
the width of pic inches [default 3]
参数说明:
-m 指定metadata文件
-v 指定要比较的变量所在的列列名,可以多个
-g 指定分组列列名
-G 指定输出 boxplot类型
使用举例:
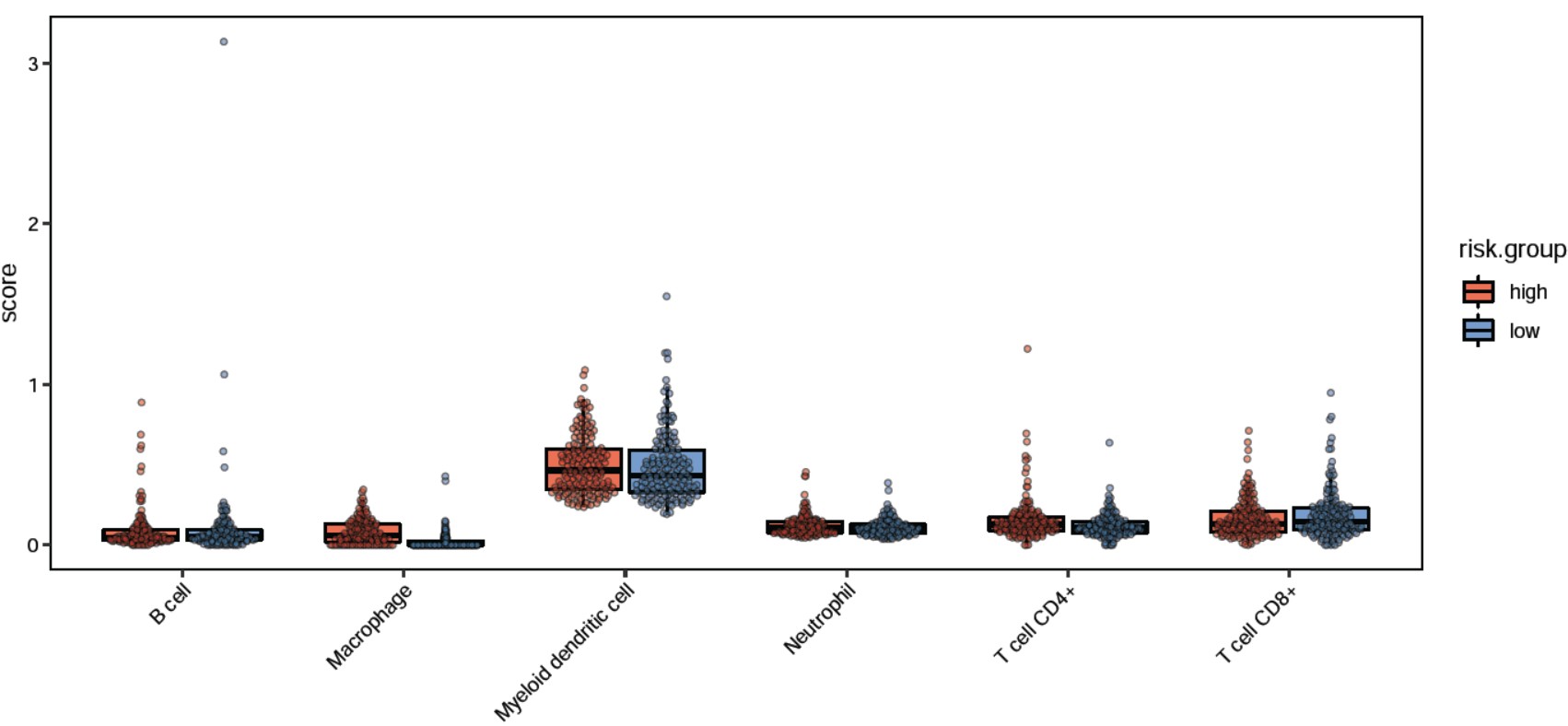
Rscript $scriptdir/compare_stat_boxplot.r -m ../08.Nomogram/nomogram_metadata.tsv \
-v PDGFRL CXCR4 PAK3 CSF1R PDCD1 \
-g risk.group -n signature.geneExp -W 10 -G boxplot -y "log2(TPM)"
结果输出:
脚本获取与使用课程:https://study.163.com/course/introduction/1211864801.htm?share=1&shareId=1030291076
- 发表于 2021-08-30 17:54
- 阅读 ( 2445 )
- 分类:TCGA
你可能感兴趣的文章
相关问题
- 请教一下TCGA的蛋白数据怎么理解,它是有正负值的,这个正负值代表什么意思,为什么蛋白表达会有负值?我只想给它分成高表达,低表达和表达缺失组应该怎么处理呀? 0 回答
- TCGA下载 docker报错 0 回答
- TCGA差异分析如何根据临床信息分组? 1 回答
- TCGA用R数据下载错误 1 回答
- 老师,想请教一下TCGA基因表达数据的问题,我从xena.ucsc网页上下载了基因表达数据TCGA-CESC.htseq_counts.tsv;然后发现该数据中只有Ensembl格式的基因ID ,没有SYMBOL格式的。所以接下来进行基因ID格式转换,却发现同一个SYMBOL ID对应的多个Ensembl格式的ID,想问下老师,这种情况该怎么处理?同一个SYMBOL ID所对应的多个Ensembl格式ID的基因表达数据应该留下哪一个? 1 回答
- TCGA的RNA-seq count annotation过滤,有些不知道是否需要删除 0 回答
1 条评论
请先 登录 后评论
