拟南芥叶单细胞分析流程
拟南芥单细胞分析流程

数据介绍:拟南芥叶处理DC3000和对照control两个样本:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE213622

数据下载
这里我们下载圆圈2的数据
wget -c https://ftp.ncbi.nlm.nih.gov/geo/series/GSE213nnn/GSE213622/suppl/GSE213622%5Fcoaker%5Fatpst%5Fsinglecell%5Fmetadata.csv.gz
wget -c https://ftp.ncbi.nlm.nih.gov/geo/series/GSE213nnn/GSE213622/suppl/GSE213622%5Fcoaker%5Fatpst%5Fsinglecell%5Fbarcodes.tsv.gz
wget -c https://ftp.ncbi.nlm.nih.gov/geo/series/GSE213nnn/GSE213622/suppl/GSE213622%5Fcoaker%5Fatpst%5Fsinglecell%5Ffeatures.tsv.gz
wget -c https://ftp.ncbi.nlm.nih.gov/geo/series/GSE213nnn/GSE213622/suppl/GSE213622%5Fcoaker%5Fatpst%5Fsinglecell%5Fmatrix.mtx.gz
#链接数据,10X文件名格式方便读入数据 ln -s GSE213622_coaker_atpst_singlecell_barcodes.tsv.gz barcodes.tsv.gz
ln -s GSE213622_coaker_atpst_singlecell_features.tsv.gz features.tsv.gz
ln -s GSE213622_coaker_atpst_singlecell_matrix.mtx.gz matrix.mtx.gz
#细胞注释文件逗号换成tab,方便后续读入数据
zcat GSE213622_coaker_atpst_singlecell_metadata.csv.gz |sed 's/,/\t/g' >meta.txt
数据转入质控:
Rscript $scripts/seurat_sc_qc.r \
-d ./ \
-p ara_leaf --project ara_leaf \
--nUMI.min 500 \ --gene.column 1 \
--nGene.min 250 \
--mito.gene.pattern "^ATM.*" \
--percent_mito 6 \
--log10GenesPerUMI 0.8 \
--metadata meta.txt
分群聚类:
Rscript $scripts/seurat_sc_cluster.r --cpu 10 --rds ara_leaf.afterQC.qs \
--sctransform \
--integrate.method harmony --batch.id Sample_Name \
--resolution 0.5 -d 30 \
-p leaf.harmony.sctransform -o leaf.harmony

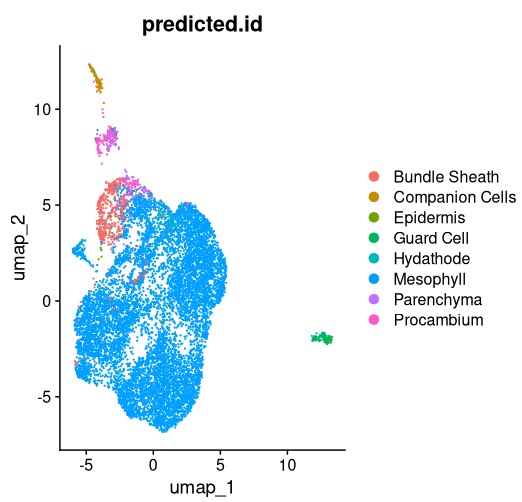
原文:https://www.biorxiv.org/content/10.1101/2022.10.07.511353v2.full
单细胞转录组数据分析实操
- 发表于 2025-01-23 14:41
- 阅读 ( 479 )
- 分类:转录组
你可能感兴趣的文章
- 单细胞转录组镜像代码更新4.0 1130 浏览
- 单细胞转录组数据挖掘流程记录-头颈癌(HNSCC)(GSE181919) 860 浏览
- 非负矩阵分解(cNMF) 1675 浏览
- 单细胞转录组如何将UCell CytoTRACE2 infercnv等结果添加到总的rds中 879 浏览
- 单细胞转录组-infer CNV拷贝数变异分析介绍 1704 浏览
0 条评论
请先 登录 后评论
