immune_infiltrates_rnaseq.r RNA-seq 转录组数据免疫侵润分析
使用方法:
$ Rscript immune_infiltrates_rnaseq.r -h
usage: immune_infiltrates_rnaseq.r
[-h] -i expset [-g gene.info] [-t type] [--tpm] [-o outdir]
RNA seq 免疫侵润分析. In general values should be TPM-normalized ,not log-
transformed.
optional arguments:
-h, --help show this help message and exit
-i expset, --expset expset
input gene expression set matrix from RNA-seq data tsv
format [required]
-g gene.info, --gene.info gene.info
input gene info data [required]
-t type, --type type TIMER uses indication-specific reference profiles.
[optional]
--tpm whether convert fpkm to tpm [optional, default: False]
-o outdir, --outdir outdir
output file directory [default cwd]
参数说明:
-i 输入基因表达量文件,建议用TPM标准化之后的数据,如果是FPKM可以设置--tpm进行转换;第一列ID为基因NAME
| ID | TCGA-D7-A74A-01A-11R-A32D-31 | TCGA-BR-7704-01A-11R-2055-13 | TCGA-VQ-A91N-01A-11R-A414-31 | TCGA-CD-A4MH-01A-11R-A251-31 |
| NUP50 | 18.65505 | 31.59232 | 28.23382 | 28.76485 |
| CXCR4 | 64.85805 | 125.123 | 56.35244 | 69.98976 |
| NT5E | 111.4818 | 69.8587 | 79.37382 | 25.05824 |
| EFNA3 | 8.247857 | 42.03308 | 43.46432 | 26.66024 |
| STC1 | 4.781111 | 21.36327 | 40.81077 | 19.51568 |
| ZBTB7A | 95.51678 | 103.4768 | 158.3024 | 126.2677 |
| CLDN9 | 1.187456 | 2.476138 | 0.366081 | 7.347344 |
-t 指定癌症类型, TIMER计算需要,可以指定类型,如果不指定 不输出TIMER的分析结果:
# "kich" "blca" "brca" "cesc" "gbm" "hnsc" "kirp" "lgg" "lihc" "luad"
# "lusc" "prad" "sarc" "pcpg" "paad" "tgct" "ucec" "ov" "skcm" "dlbc"
# "kirc" "acc" "meso" "thca" "uvm" "ucs" "thym" "esca" "stad" "read"
# "coad" "chol"
方法说明:
使用R包:immunedeconv
可以输出以下方法的免疫侵润分析结果,mcp_counter 需要联网才可以运行,有时报错不出结果;
quantiseq timer cibersort cibersort_abs mcp_counter xcell epic
不同方法文献引用:https://github.com/icbi-lab/immunedeconv
| quanTIseq | free (BSD) | Finotello, F., Mayer, C., Plattner, C., Laschober, G., Rieder, D., Hackl, H., ..., Sopper, S. (2019). Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome medicine, 11(1), 34. https://doi.org/10.1186/s13073-019-0638-6 |
| TIMER | free (GPL 2.0) | Li, B., Severson, E., Pignon, J.-C., Zhao, H., Li, T., Novak, J., … Liu, X. S. (2016). Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biology, 17(1), 174. https://doi.org/10.1186/s13059-016-1028-7 |
| CIBERSORT | free for non-commerical use only | Newman, A. M., Liu, C. L., Green, M. R., Gentles, A. J., Feng, W., Xu, Y., … Alizadeh, A. A. (2015). Robust enumeration of cell subsets from tissue expression profiles. Nature Methods, 12(5), 453–457. https://doi.org/10.1038/nmeth.3337 |
| MCPCounter | free (GPL 3.0) | Becht, E., Giraldo, N. A., Lacroix, L., Buttard, B., Elarouci, N., Petitprez, F., … de Reyniès, A. (2016). Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biology, 17(1), 218. https://doi.org/10.1186/s13059-016-1070-5 |
| xCell | free (GPL 3.0) | Aran, D., Hu, Z., & Butte, A. J. (2017). xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biology, 18(1), 220. https://doi.org/10.1186/s13059-017-1349-1 |
| EPIC | free for non-commercial use only (Academic License) | Racle, J., de Jonge, K., Baumgaertner, P., Speiser, D. E., & Gfeller, D. (2017). Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. ELife, 6, e26476. https://doi.org/10.7554/eLife.26476 |
免疫侵润分析方法
- 基于标记基因的方法 a
- 基于反卷积的方法 b
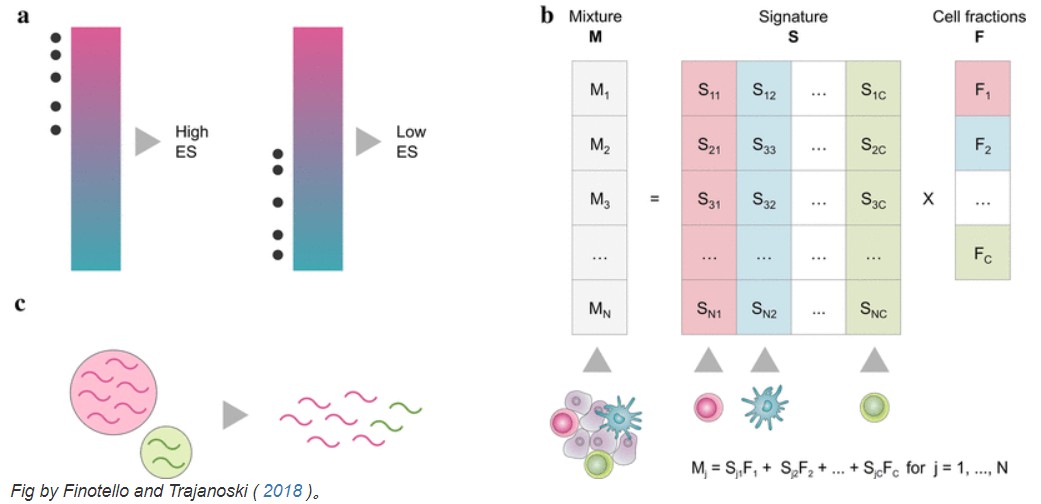
(a)基于标记基因的方法: 基于基因列表(signature),代表细胞类型的特征。通过查看特征基因的表达值,可以独立量化每种细胞类型,直接使用基因表达值(MCP-counter)或通过执行特征富集的统计测试(xCell)。
(b)反卷积方法: 将问题表述为一个方程组,该方程组将样本的基因表达描述为不同细胞类型贡献的加权总和。通过求解逆问题,可以在给定特征矩阵和混合基因表达的情况下推断细胞类型分数。如 ν-Support Vector Regression (SVR) (CIBERSORT) ,约束最小二乘回归 (quantTIseq, EPIC) 或线性最小二乘回归 (TIMER)。
有关更多信息,请查看Finotello 和 Trajanoski ( 2018 ) 的综述。
不同软件计算免疫得分scores解读
In general, cell-type scores allow for the comparison (1) between samples, (2) between cell-types or (3) both. Between-sample comparisons allow to make statements such as “In patient A, there are more CD8+ T cells than in patient B”. Between-cell-type comparisons allow to make statements such as “In a certain patient, there are more B cells than T cells”. For more information, see our Benchmark paper (Sturm et al. (2019)).
Methods that allow between-sample comparisons
MCP-counter xCell TIMER
Methods that allow between-cell-type comparisions
CIBERSORT
Methods that allow both
EPIC quanTIseq
- 发表于 2021-06-15 16:18
- 阅读 ( 3390 )
- 分类:TCGA
你可能感兴趣的文章
- 胃癌免疫侵润预后Signature数据挖掘-糖酵解 4592 浏览
- 胃癌免疫侵润预后Signature数据挖掘-4.8分SCI思路解析 5955 浏览
- tsne_analysis.r 表达聚类分析 3237 浏览
- immune_box_plot.r 免疫侵润box分布图 2894 浏览
- immune_compare_stat.r 免疫细胞表达差异比较 2487 浏览
- immune_bar_plot.r 免疫侵润分析bar绘图 2489 浏览
