immune_box_plot.r 免疫侵润box分布图
immune_box_plot.r 免疫侵润box分布图
使用方法
$ Rscript $scriptdir/immune_box_plot.r -h
usage: immune_box_plot.r [-h] -i filepath -m
filepath -g group
[-b groupby]
[-p palette]
[-G geom [geom ...]]
[-T title] [-x xlab]
[-y ylab] [-o path]
[-n prefix] [-H number]
[-W number]
immune box plot : https://www.omicsclass.com/article/1497
optional arguments:
-h, --help show this help message and exit
-i filepath, --input filepath
input immune result file[required]
-m filepath, --metadata filepath
input metadata file path[required]
-g group, --group group
group name from metadata to test[required]
-b groupby, --groupby groupby
main group name from metadata to subset [default NULL]
-p palette, --palette palette
fill palette in ggsci : eg npg lancet... for more
info:https://nanx.me/ggsci/articles/ggsci.html
[default lancet]
-G geom [geom ...], --geom geom [geom ...]
set type of plot: boxplot, point, violin, splitviolin
[default=boxplot]
-T title, --title title
the label for main title [optional, default: 'imm']
-x xlab, --xlab xlab input xlab [default cell type]
-y ylab, --ylab ylab input ylab [default fraction]
-o path, --outdir path
output file directory [default cwd]
-n prefix, --name prefix
out file name prefix [default demo]
-H number, --height number
the height of pic inches [default 5]
-W number, --width number
the width of pic inches [default 8]
使用命令举例
#box图展示
Rscript $scriptdir/immune_box_plot.r -i immu/ssgsea.res.tsv \
-m metadata.group.tsv -g subtype.hclust -y NES \
-G boxplot -o immu -n ssgse
#splitviolin图展示
Rscript $scriptdir/immune_box_plot.r -i immu/ssgsea.res.tsv \
-m metadata.group.tsv -g subtype.hclust -y NES \
-G splitviolin -o immu -n ssgse
绘图结果:
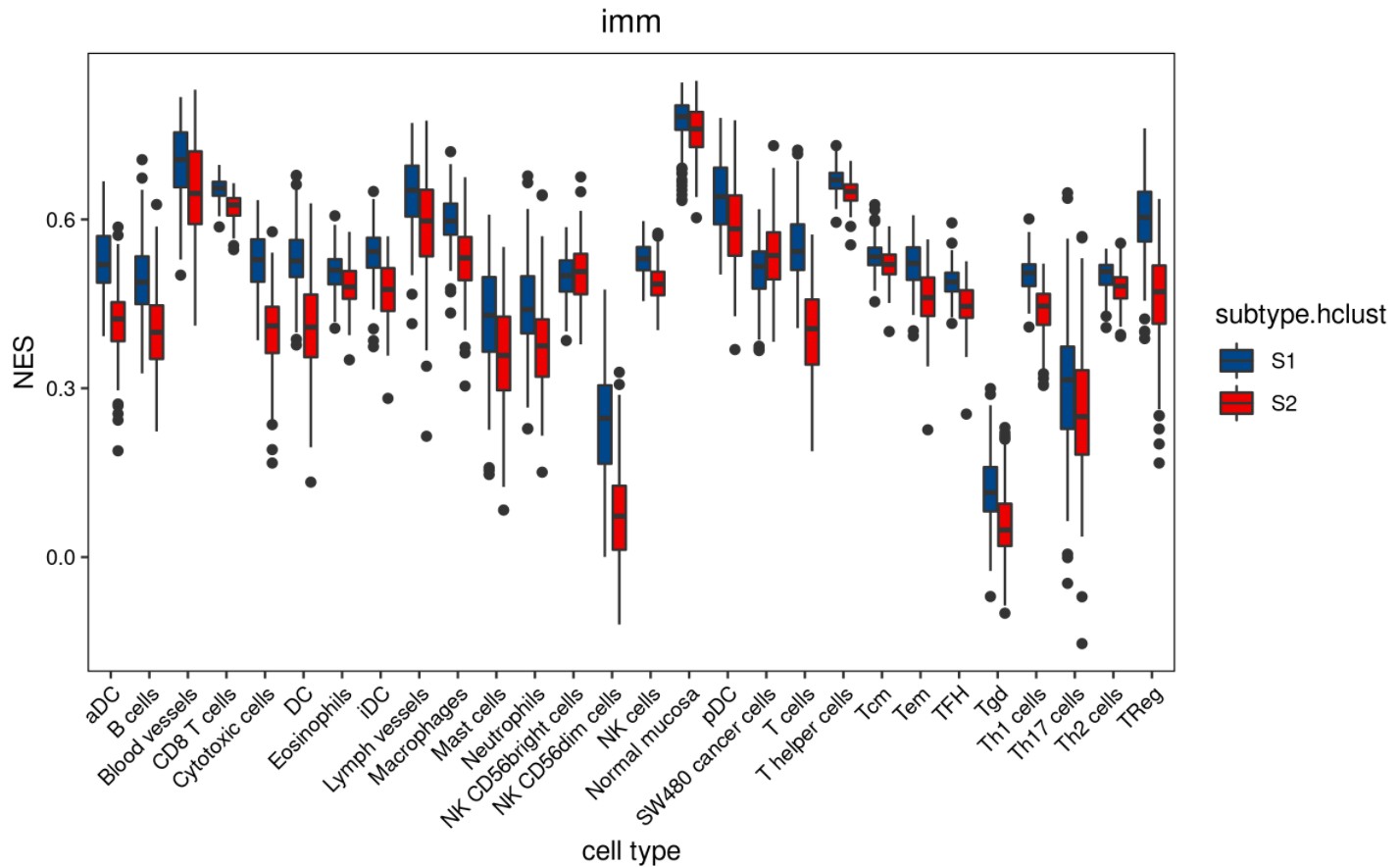
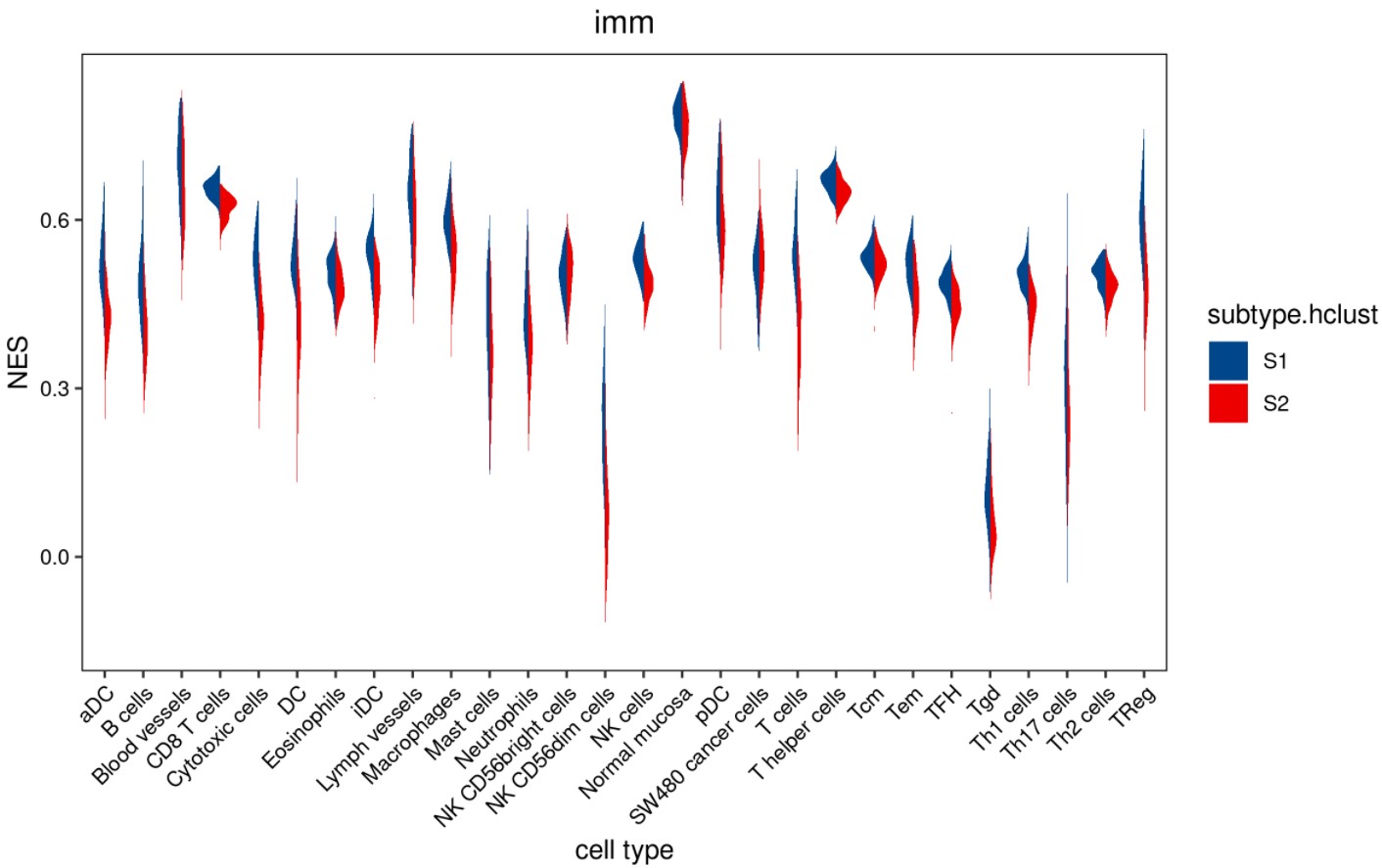
- 发表于 2021-06-22 12:11
- 阅读 ( 2517 )
- 分类:TCGA
你可能感兴趣的文章
- 胃癌免疫侵润预后Signature数据挖掘-糖酵解 3800 浏览
- 胃癌免疫侵润预后Signature数据挖掘-4.8分SCI思路解析 5204 浏览
- tsne_analysis.r 表达聚类分析 2794 浏览
- immune_compare_stat.r 免疫细胞表达差异比较 2108 浏览
- immune_bar_plot.r 免疫侵润分析bar绘图 2152 浏览
0 条评论
请先 登录 后评论
