tsne_analysis.r 表达聚类分析
tsne_analysis.r 表达聚类分析
使用说明:
$ Rscript $scriptdir/tsne_analysis.r -h
usage: /work/my_stad_immu/scripts/tsne_analysis.r [-h] -i filepath -m metadata
[-d dims] [-p perplexity]
[--theta theta]
[-M initial_dims] [-T top]
-g group [-s size]
[-a alpha] [-e] [-L]
[-X x.lab] [-Y y.lab]
[-t title] [-o outdir]
[-n prefix] [-H height]
[-W width]
t-Distributed Stochastic Neighbor Embedding t-SNE analysis:
https://www.omicsclass.com/article/1498
optional arguments:
-h, --help show this help message and exit
-i filepath, --input filepath
input the dataset martix [required]
-m metadata, --metadata metadata
input metadata file path[required]
-d dims, --dims dims set Output dimensionality [default=2]
-p perplexity, --perplexity perplexity
Perplexity parameter (should not be bigger than 3 *
perplexity < nrow(X) - 1, see details for
interpretation) [default 50]
--theta theta Speed/accuracy trade-off (increase for less accuracy),
set to 0.0 for exact TSNE [default 0.5]
-M initial_dims, --initial_dims initial_dims
the number of dimensions that should be retained in
the initial PCA step [default 50]
-T top, --top top select top gene to analysis [default NULL]
-g group, --group group
input group id in metadata file to fill
color[required]
-s size, --size size point size [optional, default: 3]
-a alpha, --alpha alpha
point transparency [0-1] [optional, default: 1]
-e, --ellipse whether draw ellipse [optional, default: False]
-L, --label whether show pionts sample name [optional, default:
False]
-X x.lab, --x.lab x.lab
the label for x axis [optional, default: t-SNE1]
-Y y.lab, --y.lab y.lab
the label for y axis [optional, default: t-SNE2]
-t title, --title title
the label for main title [optional, default: t-SNE]
-o outdir, --outdir outdir
output file directory [default cwd]
-n prefix, --name prefix
out file name prefix [default demo]
-H height, --height height
the height of pic inches [default 5]
-W width, --width width
the width of pic inches [default 5]
参数说明
| dims | 参数设置降维之后的维度,默认是2 |
| perplexity | 困惑度,参数须取值小于 (样本数量-1)/3 |
| theta | 参数越大,结果的准确度越低,默认是0.5 |
| max_iter | 最大迭代次数 |
| pca | 表示是否对输入的原始数据进行PCA分析,然后用分析后的数据进行后续分析,默认TRUE |
使用举例:
#降维验证分组
#####t-SNE
Rscript $scriptdir/tsne_analysis.r -i immu/ssgsea.res.tsv -m metadata.group.tsv \
-o tsne -g subtype.hclust -p 6 -n tsne
结果展示:
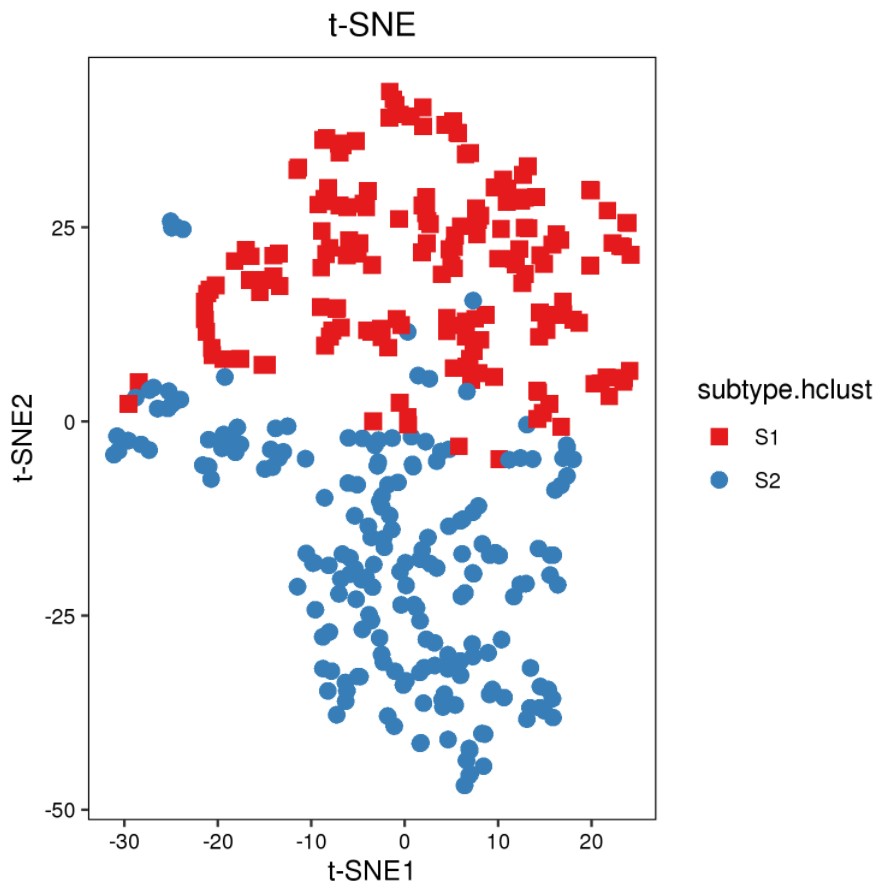 引用:
引用:
Rtsne包 https://github.com/jkrijthe/Rtsne
- 发表于 2021-06-22 12:20
- 阅读 ( 2795 )
- 分类:TCGA
你可能感兴趣的文章
- 胃癌免疫侵润预后Signature数据挖掘-糖酵解 3801 浏览
- 胃癌免疫侵润预后Signature数据挖掘-4.8分SCI思路解析 5204 浏览
- immune_box_plot.r 免疫侵润box分布图 2517 浏览
- immune_compare_stat.r 免疫细胞表达差异比较 2108 浏览
- immune_bar_plot.r 免疫侵润分析bar绘图 2152 浏览
0 条评论
请先 登录 后评论
