enrichKEGG_pip.r KEGG 富集分析
enrichKEGG_pip.r KEGG 富集分析
使用说明:
$Rscript $scriptdir/enrichKEGG_pip.r -h
usage: /work/my_stad_immu/scripts/enrichKEGG_pip.r [-h] -g gene.list
[-d ann.db] [-t idtype]
[-s show.type]
[-O organism]
[-p pvalueCutoff]
[-q qvalueCutoff]
[-c showCategory]
[-n prefix] [-o outdir]
[-H height] [-W width]
KEGG enrich analysis : https://www.omicsclass.com/article/1503
optional arguments:
-h, --help show this help message and exit
-g gene.list, --gene.list gene.list
diff expressed gene list file, required
-d ann.db, --ann.db ann.db
org.Hs.eg.db or org.Mm.eg.db ,for more info visit:
https://www.omicsclass.com/article/1244 [optional,
default: org.Hs.eg.db ]
-t idtype, --idtype idtype
deg gene id type: ENSEMBL SYMBOL ENTREZID GENENAME
[optional, default: SYMBOL ]
-s show.type, --show.type show.type
set example ID type name [optional, default: ENTREZID
]
-O organism, --organism organism
organism ,for more info visit:
https://www.omicsclass.com/article/787 [optional,
default: hsa ]
-p pvalueCutoff, --pvalueCutoff pvalueCutoff
pvalue cutoff on enrichment tests to report,
[optional, default: 0.05 ]
-q qvalueCutoff, --qvalueCutoff qvalueCutoff
qvalue cutoff on enrichment tests to report as
significant. Tests must pass i) pvalueCutoff on
unadjusted pvalues, ii) pvalueCutoff on adjusted
pvalues and iii) qvalueCutoff on qvalues to be
reported., [optional, default: 0.2 ]
-c showCategory, --showCategory showCategory
how many KEGG Category to show, [optional, default: 10
]
-n prefix, --prefix prefix
the output file prefix [optional, default: KEGG ]
-o outdir, --outdir outdir
output file directory [default cwd]
-H height, --height height
the height of pic inches [default 5]
-W width, --width width
the width of pic inches [default 7]
参数说明:
-g 输入基因列表文件: 脚本会读取第一列基因ID作为基因集:
-d 物种注释数据库: 人的:org.Hs.eg.db
-t 指定输入基因列表的ID类型
使用举例:
Rscript $scriptdir/enrichKEGG_pip.r --gene.list $workdir/04.deg/S1_vs_S2.DEG.final.tsv \
-o KEGG -n S1_vs_S2 --pvalueCutoff 0.01 --ann.db org.Hs.eg.db --organism hsa \
--idtype SYMBOL
结果展示:
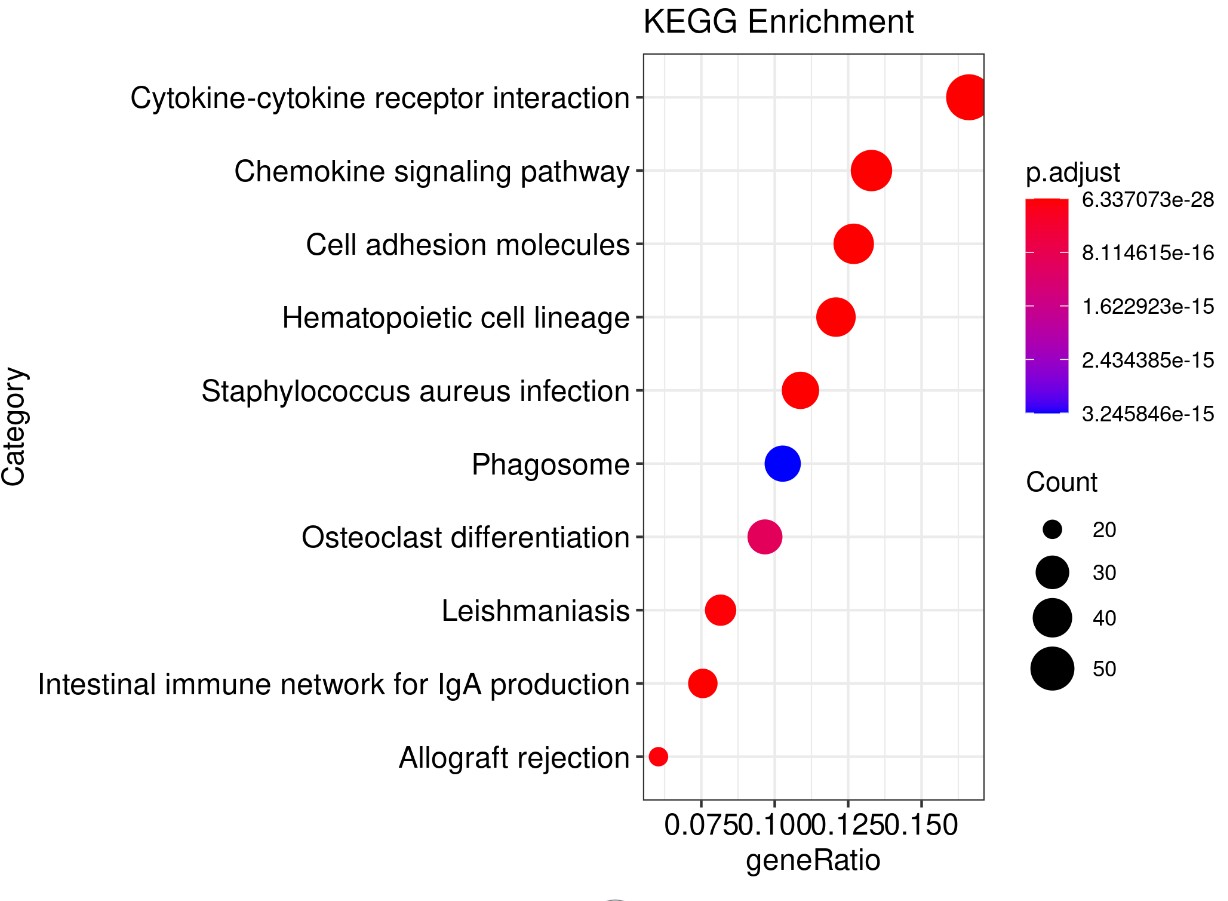
参考文献:
Yu G, Wang L, Han Y, He Q (2012). “clusterProfiler: an R package for comparing biological themes among gene clusters.” OMICS: A Journal of Integrative Biology, 16(5), 284-287. doi: 10.1089/omi.2011.0118.
- 发表于 2021-06-23 13:37
- 阅读 ( 3788 )
- 分类:转录组
你可能感兴趣的文章
- metascape数据库 7496 浏览
- eggNOG批量注释基因组数据库 4764 浏览
- clusterProfiler非模式物种 KEGG与GO富集分析 13939 浏览
- enrichGSEA_pip.r GSEA富集分析 3350 浏览
- enrichGO_pip.r GO富集分析 3257 浏览
0 条评论
请先 登录 后评论
