nomogram_multi_cox.r 多因素cox分析构建模型并绘制列线图nomogram
nomogram_multi_cox.r 多因素cox分析构建模型并绘制列线图nomogram
使用说明:
使用方法类似于基因表达量多因素cox分析https://www.omicsclass.com/article/1533 ,但这个脚本支持输入临床数据用于模型构建并绘制列线图,列线图用的R包rms;
$Rscript $scriptdir/nomogram_multi_cox.r -h
usage: /share/nas1/huangls/test/TCGA_immu/scripts/nomogram_multi_cox.r
[-h] -i data -t time -e event -v variate [variate ...]
[-P predict.time [predict.time ...]] [-c cut.score] [-s seed]
[-o outdir] [-p prefix]
cox regression analysis gene expression
optional arguments:
-h, --help show this help message and exit
-i data, --data data input data file path[required]
-t time, --time time set suvival time column name [required]
-e event, --event event
set event column name must 0 or 1 code format
[required]
-v variate [variate ...], --variate variate [variate ...]
variate for cox analysis [required]
-P predict.time [predict.time ...], --predict.time predict.time [predict.time ...]
Time point to draw the ROC curve [default 365 1095
1825]
-c cut.score, --cut.score cut.score
set cut score value to divide high and low risk groups
[default median]
-s seed, --seed seed set random seed [default 2021]
-o outdir, --outdir outdir
output file directory [default cwd]
-p prefix, --prefix prefix
out file name prefix [default cox]
使用举例:
Rscript $scriptdir/nomogram_multi_cox.r -i nomogram_metadata.tsv -e EVENT -t TIME \
-v riskScore ajcc_pathologic_stage -o multicox.nomogram\
-p multicox -P 365 710 1095
结果展示:
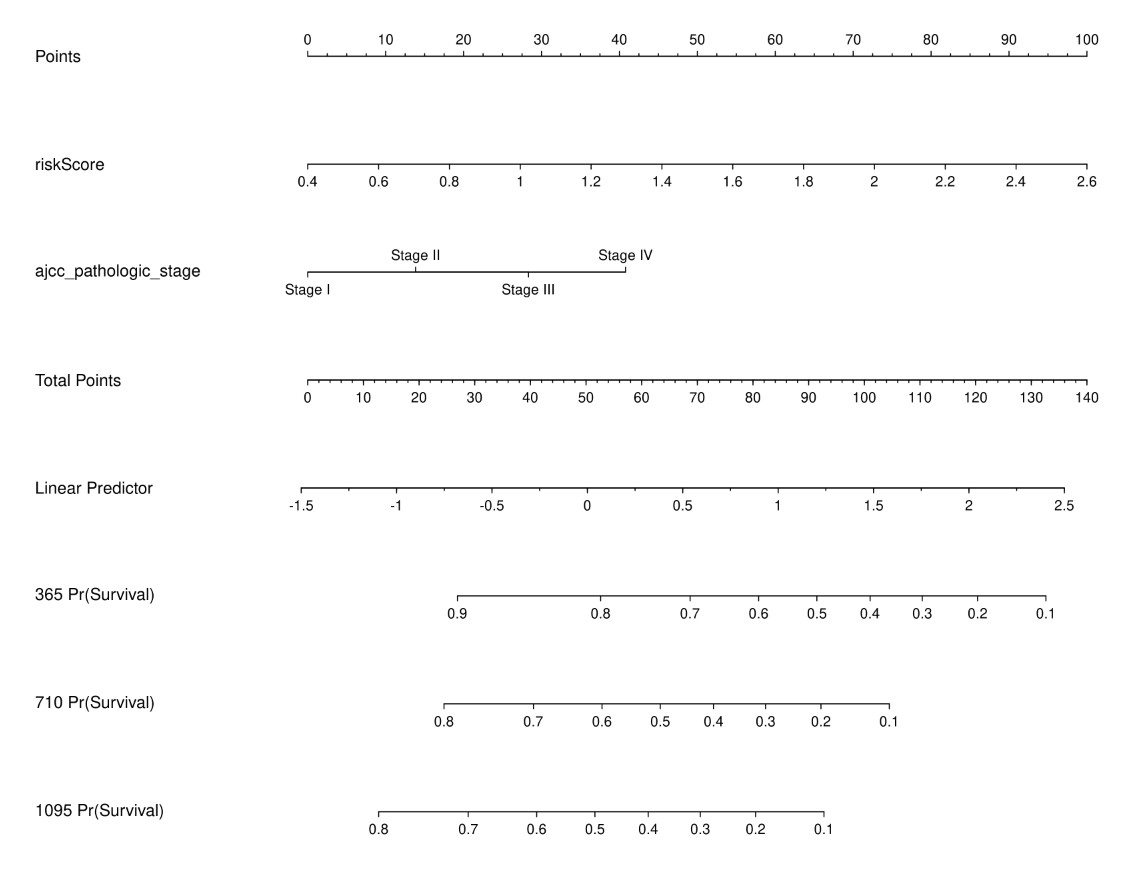 脚本获取与使用课程:https://study.163.com/course/introduction/1211864801.htm?share=1&shareId=1030291076
脚本获取与使用课程:https://study.163.com/course/introduction/1211864801.htm?share=1&shareId=1030291076
- 发表于 2021-08-30 17:25
- 阅读 ( 2688 )
- 分类:临床医学
你可能感兴趣的文章
相关问题
0 条评论
请先 登录 后评论
