vcf文件annovar的注释结果绘制瀑布图maftools分析
vcf文件annovar的注释结果绘制瀑布图maftools分析
1.vcf文件annovar注释:
table_annovar.pl 154.raw.somatic.vcf.gz humandb/hg38/ -buildver hg38 -out 154 -remove -protocol \
refGene,cosmic70,nci60,esp6500siv2_all,clinvar_20210501,1000g2015aug_all,1000g2015aug_eas,1000g2015aug_sas,avsnp150,gwasCatalog,ljb26_all,cytoBand,dgvMerged,phastConsElements100way,genomicSuperDups -operation g,f,f,f,f,f,f,f,f,f,f,r,r,r,r -nastring . -vcfinput
2. 提取必要的信息
for i in *.hg38_multianno.txt
do
sample=`echo $i|awk -F '.' '{print $2}'`
cut -f '1-10' $i|sed '1d'|sed "s/$/\t${sample}/">>all_sample.txt
done
sed -i '1s/^/Chr\tStart\tEnd\tRef\tAlt\tFunc.refGene\tGene.refGene\tGeneDetail.refGene\tExonicFunc.refGene\tAAChange.refGene\tTumor_Sample_Barcode\n/' all_sample.txt
3. 读入数据,利用maftools绘图
library(maftools) var_maf= annovarToMaf(annovar = "all_sample.txt",
Center = 'NA', refBuild = 'hg38', tsbCol = 'Tumor_Sample_Barcode', table = 'refGene',MAFobj =T, sep = "\t") plotmafSummary(maf = var_maf, rmOutlier = TRUE, addStat = 'median')
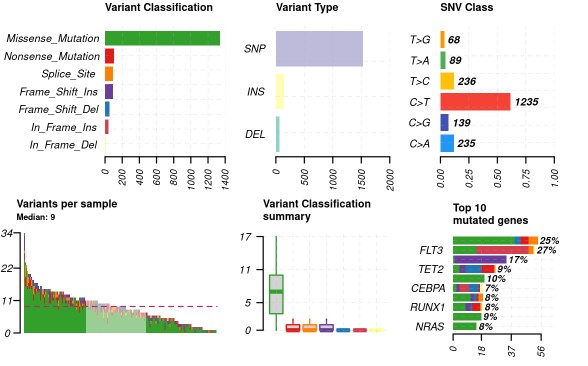
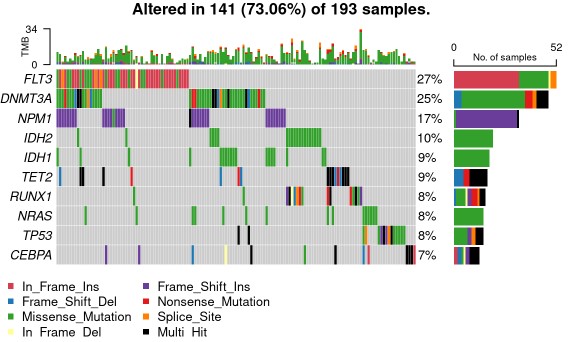
oncoplot(maf = var_maf, top = 30, fontSize = 12 ,showTumorSampleBarcodes = F )
- 发表于 2022-01-04 13:04
- 阅读 ( 4466 )
- 分类:重测序
你可能感兴趣的文章
- TCGA SNV 体细胞突变下载 2678 浏览
- maftools|突变数据可视化-瀑布图绘制及脚本使用 12122 浏览
- maftools 癌症组织突变频谱展示工具 3956 浏览
相关问题
0 条评论
请先 登录 后评论
