RagTag:同源锚定延伸
RagTag是一个集合功能的软件可以用来构建scaffold、提升基因组组装质量
RagTag是一个集合功能的软件可以用来构建scaffold、提升基因组组装质量,其主要功能用法如下:
usage: ragtag.py <command> [options]
assembly improvement:
correct homology-based misassembly correction
scaffold homology-based assembly scaffolding
patch homology-based assembly patching
merge scaffold merging
file utilities:
agp2fa build a FASTA file from an AGP file
agpcheck check for valid AGP file format
asmstats assembly statistics
splitasm split an assembly at gaps
delta2paf delta to PAF file conversion
paf2delta PAF to delta file conversion
updategff update gff intervals
options:
-c, --citation
-v, --version
一、组装
RagTag的组装流程一共四步:correct,scaffold,patch,merge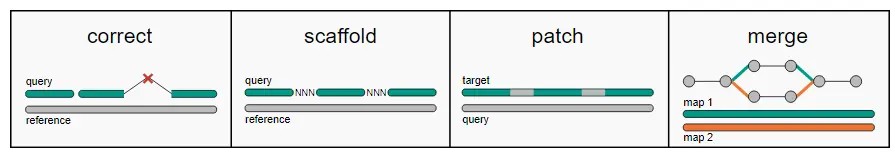
1. correct 错配校正
校正是使用参考基因组来鉴定和校正contigs中的组装错误,该步骤不会将序列减少或增加,仅仅是将序列在错误组装的位置进行打断。
ragtag.py correct <reference.fa> <query.fa>
2. scaffold
将相邻的contigs序列用100个N连起来,序列的位置和方向需要根据与参考基因组的比对结果确定。
ragtag.py scaffold <reference.fa> <query.fa>
#添加-C会将未组装上的序列连成chr0
3. patch
用contigs序列对上一步得到的scaffold序列进行gap填补。
ragtag.py patch <target.fa> <query.fa>
4. merge
在scaffolding过程中,可能会根据不同参数或图谱数据产生多个版本的基因组组装结果,该步骤可以将多个结果根据权重进行最终组装结果的生成。
ragtag.py merge <asm.fa> <scf1.agp> <scf2.agp> [...]
<asm.fasta> assembly fasta file (uncompressed or bgzipped)
<scf1.agp> <scf2.agp> [...]
scaffolding AGP files
二、文件处理
除组装之外,RagTag还有其他功能,具体如最上方代码块所示,如对组装结果进行统计
# 得到N50
ragtag.py asmstats fa文件
n sum genome_size min max auN N50 N90 L50 L90 file 709 157665521 NA 13698 26961813 11328043 12446067 38225 5 139 fa文件路径
#得到NG50
ragtag.py asmstats -g 130000000 fa文件
n sum genome_size min max auN NG50 NG90 LG50 LG90 file
709 157665521 130000000 13698 26961813 13738783 13410975 5073862 4 10 fa文件
参考:
- 发表于 2023-10-25 13:35
- 阅读 ( 1833 )
- 分类:软件工具
你可能感兴趣的文章
相关问题
0 条评论
请先 登录 后评论
