叶绿体或线粒体重复序列圈图绘制
叶绿体或线粒体重复序列圈图绘制
细胞器基因组分析的重复序列展示中有时会展示为下面这种circos圈,用来描述基因组的微卫星重复序列(SSR)、串联重复序列(Tandem repeats)的分布以及散在重复序列(Dispersed repeats)的共线性关系,简洁美观,今天带大家一起来画一下细胞器重复序列的圈图。
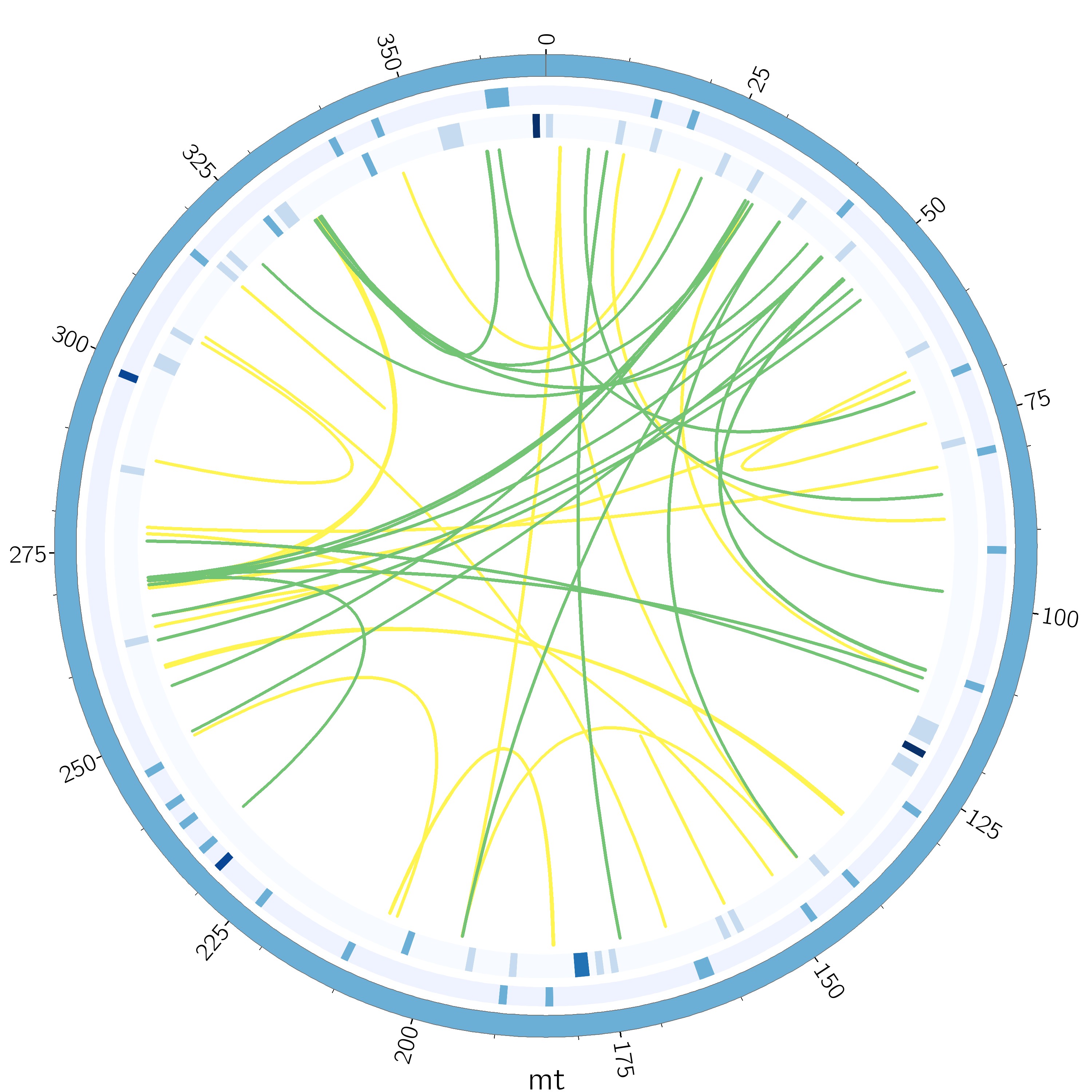
一、重复序列鉴定及整理
以下为鉴定细胞器基因组的SSR、串联重复序列和散在重复序列的教程
Tandem repeats:https://www.omicsclass.com/article/2363
Dispersed repeats:https://www.omicsclass.com/article/2298
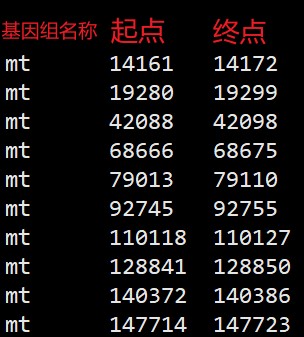
将SSR、串联重复序列的结果整理成如下格式:
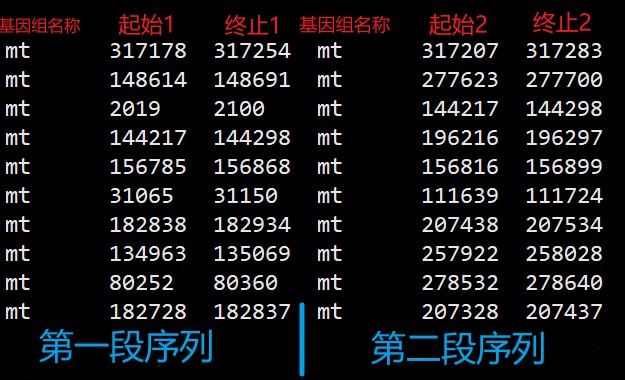
散在重复序列的结果,需自行计算终止位置,且按不同类型 Forward(F), Palindromic (P) 整理成如下格式,注意每个类型单独整理,分别命名为link.f.txt和link.p.txt文件。
二、输入文件准备
1.染色体文件 karyotype.txt 其中第1、2、5列是固定的,关于颜色可直接指定,也可用色板

2.SSR及TRF文件
#首先准备染色体长度文件chr.txt如下
mt 367808
#用bedtools以500kb为滑窗,沿chr.txt中的基因组长度创建窗口
bedtools makewindows -g chr.txt -w 1000 > windows.bed
#统计信息,生成SSR及TRF绘图文件
bedtools coverage -a windows.bed -b ssr.bed | cut -f 1-4 > ssr.density.txt
bedtools coverage -a windows.bed -b trf.bed | cut -f 1-4 > trf.density.txt
3.散在重复序列共线性文件
即第二步骤中整理的正向重复link.f.txt和回文重复link.p.txt文件
三、生成circos绘图配置文件
echo "
karyotype =karyotype.txt #指定染色体文件,注意这里写的相对路径,
chromosomes_units = 1000 #设置长度单位,表示500k长度的序列代表为1u
chromosomes_display_default = yes #默认是将所有的染色体都展示出来
show_tick_labels=yes
show_ticks=yes
##ideogram 染色体和刻度线配置
<ideogram>
fill=yes
label_font=default
label_parallel=yes
label_radius=dims(image,radius)-60p
label_size=65
radius=0.9r
show_label=yes
<spacing>
default=0u #染色体之间间隙
</spacing>
stroke_color=dgrey
stroke_thickness=2p
thickness=60p
</ideogram>
<ticks> #设置染色体刻度
color=black
format=%d
multiplier=1e-3
radius=1r
thickness=2p
<tick>
size=10p
spacing=10u
</tick>
<tick>
color=black
format=%d
label_offset=10p
label_size=50p
show_label=yes
size=15p
spacing=25u
thickness=4p
</tick>
</ticks>
<links> #共线性links
<link>
file=link.f.txt
bezier_radius=0r # #设置贝塞尔曲线半径,该值设大后曲线扁平。
#bezier_radius_purity=0.75
color=yellow
#crest=0.5
radius=0.85r #设置link半径
thickness=8 #设置 link 曲线的粗细
ribbon=no #条带yes,线 no
</link>
<link>
file=link.p.txt
bezier_radius=0r # #设置贝塞尔曲线半径,该值设大后曲线扁平。
#bezier_radius_purity=0.75
color=green
#crest=0.5
radius=0.85r #设置link半径
thickness=8 #设置 link 曲线的粗细
ribbon=no #条带yes,线 no
</link>
</links>
<plots>
<plot>
type = heatmap
file = ssr.density.txt #SSR文件
color = blues-7-seq
r1 = 0.98r
r0 = 0.94r
</plot>
<plot>
type = heatmap
file = trf.density.txt #TRF文件
color = blues-9-seq
r1 = 0.92r
r0 = 0.87r
</plot>
</plots>
<colors>
<<include etc/colors.conf>>
<<include etc/brewer.conf>>
#<<include etc/colors_fonts_patterns.conf>>
#<<include colors.ucsc.conf>>
#<<include colors.hsv.conf>>
</colors>
<fonts>
<<include etc/fonts.conf>>
</fonts>
<image>
<<include etc/image.conf>>
</image>
<<include etc/housekeeping.conf>>
">circos.conf
四、绘图
circos -conf circos.conf -outputfile repeat.circos
-conf:指定配置文件;-outputfile输出文件名
一般会输出png和svg两个格式
这样,一个简洁美观的重复序列圈图就完成啦,一起来试试吧~
- 发表于 2023-12-18 17:12
- 阅读 ( 4425 )
- 分类:软件工具
你可能感兴趣的文章
- 叶绿体或线粒体基因组重复序列分类及鉴定 7681 浏览
- 基因组重复序列分类 7283 浏览
相关问题
0 条评论
请先 登录 后评论
