R语言绘制差异表达火山图
R语言绘制差异表达火山图
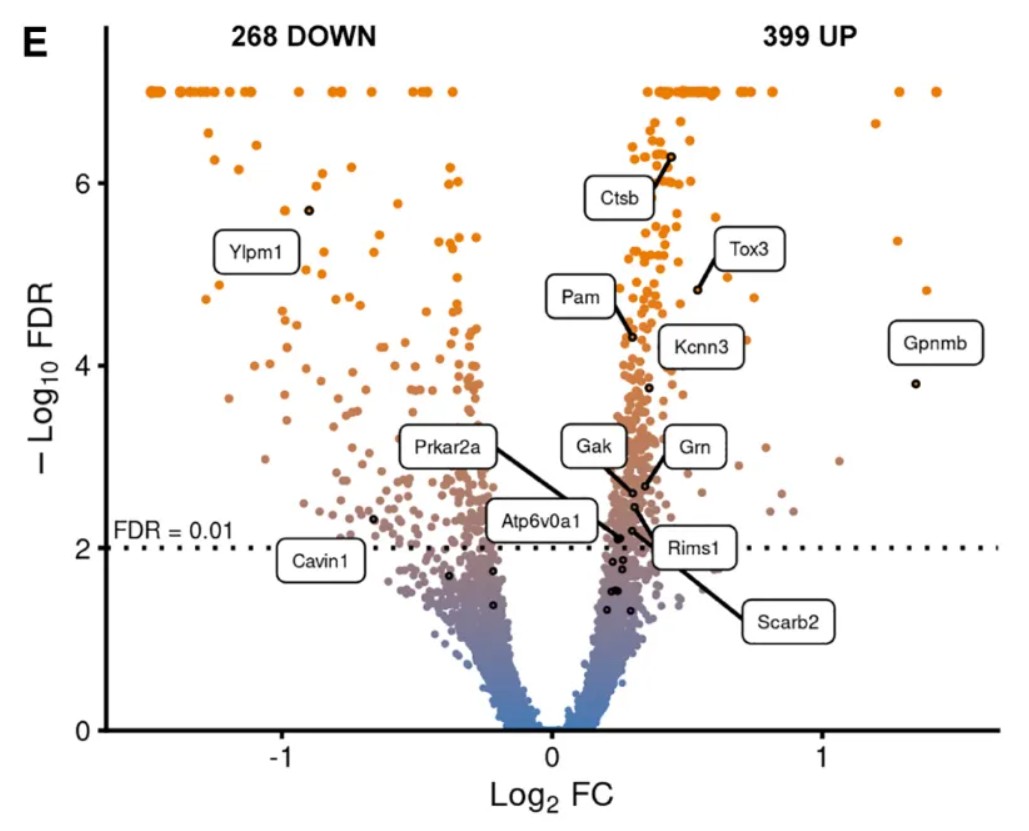
下图是一篇文献中添加了基因标签的渐变色火山图,今天教大家如何使用R语言绘制一张类似的火山图。来自文章:Cell Rep. 2024;43(3):113784.
1.安装R包
#设置镜像,
local({r <- getOption("repos")
r["CRAN"] <- "http://mirrors.tuna.tsinghua.edu.cn/CRAN/"
options(repos=r)})
# 依赖包列表:自动加载并安装
package_list <- c("ggplot2","ggrepel","dplyr","devtools")
# 判断R包加载是否成功来决定是否安装后再加载
for(p in package_list){
if(!suppressWarnings(suppressMessages(require(p, character.only = TRUE, quietly = TRUE, warn.conflicts = FALSE)))){
install.packages(p, warn.conflicts = FALSE)
suppressWarnings(suppressMessages(library(p, character.only = TRUE, quietly = TRUE, warn.conflicts = FALSE)))
}
}
2.设置FC和FDR的阈值线
fc_cutoff <- 2 fdr_cutoff <- 0.01
3.读入数据
#设置工作路径 setwd("/share/work") data<-read.table("Volcano.txt", header=TRUE,sep = "\t")
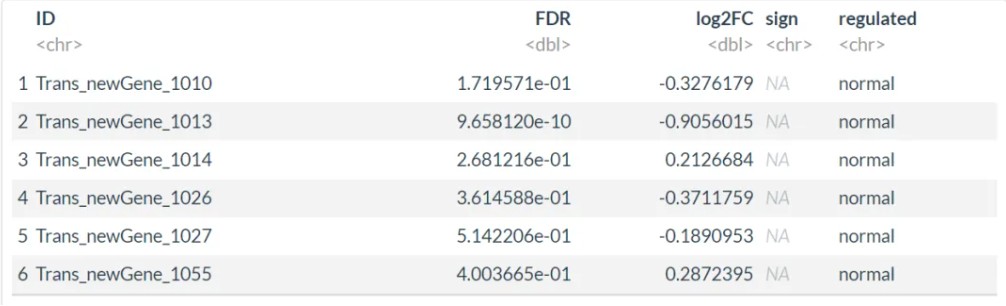
数据格式如下(表头不要改):
ID FDR log2FC sign Trans_newGene_1010 0.1719571 -0.327617885 NA Trans_newGene_1013 9.65812E-10 -0.905601526 NA Trans_newGene_1014 0.268121627 0.212668419 NA Trans_newGene_1026 0.361458835 -0.371175921 NA Trans_newGene_1027 0.514220593 -0.189095293 NA Trans_newGene_1055 0.400366453 0.287239534 NA Trans_newGene_1069 0.038485409 0.667683474 NA Trans_newGene_1071 0.65302006 0.180040874 NA Trans_newGene_1084 5.27862E-05 1.314071476 NA
第一列:基因ID;第二列:FDR值;第三列:log2FC值;第四列:要显示的基因名称,不显示的都用NA。
4.添加上下调信息列-regulated
data$regulated <- case_when(data$log2FC > log2(fc_cutoff) & data$FDR < fdr_cutoff ~ "up",
data$log2FC < -log2(fc_cutoff) & data$FDR < fdr_cutoff ~ "down",
abs(data$log2FC) <= log2(fc_cutoff) ~ "normal",
data$FDR >= fdr_cutoff ~ "normal")
head(data)
添加上下调信息后的数据:
#转换为因子 data$regulated <- factor(data$regulated, levels = c("up","down","normal"), ordered = T)
5.ggplot2绘图
mycolor1 <- c("#008EBE","#FF890E")
top.mar=0.2
right.mar=0.2
bottom.mar=0.2
left.mar=0.2
p <-ggplot(data=data,aes(log2FC,-log10(FDR),color=-log10(FDR))) +
geom_point(size=2,alpha=0.9)+
scale_colour_gradient2(low = mycolor1[1],
mid = mycolor1[2],
high = mycolor1[2],
midpoint = 50, #midpoint = 75 指定颜色渐变的中点,这意味着在数值为75时颜色会是mid指定的颜色
guide = "none") +
geom_point(data = subset(data, !is.na(sign)),
aes(x = log2FC, y = -log10(FDR)),
shape = 1, size = 2, stroke = 1.2, color = "black") + # 描边的点
geom_label_repel(aes(label=sign), fontface="bold",
color="grey50", box.padding=unit(0.35, "lines"),
point.padding=unit(0.5, "lines"), segment.colour = "grey50")+
geom_hline(yintercept = c(-log10(fdr_cutoff)),
linewidth = 0.5,
color = "orange",
lty = "dashed")+
theme_classic()+
theme(text=element_text(family = "sans",colour ="gray30",size = 10),
axis.line = element_line(linewidth = 0.6,colour = "gray30"),
axis.ticks = element_line(linewidth = 0.6,colour = "gray30"),
axis.ticks.length = unit(1.5,units = "mm"),
plot.margin=unit(x=c(top.mar,right.mar,bottom.mar,left.mar),
units="inches"))
p
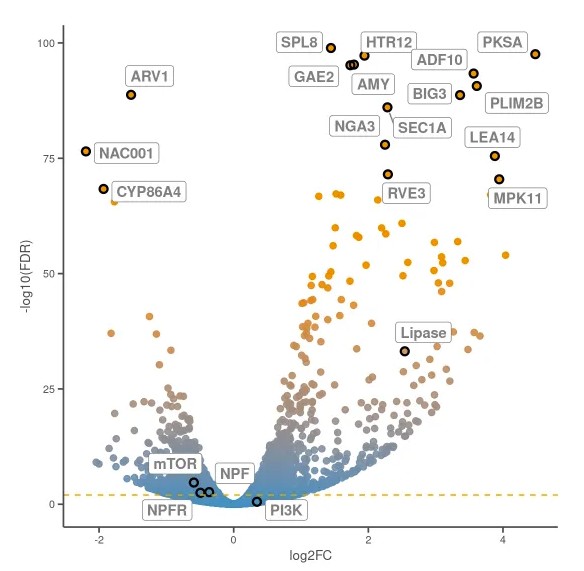
结果如下图所示:
好了,小编就先给大家介绍到这里。希望对您的科研能有所帮助!祝您工作生活顺心快乐!
参考文献:Kilfeather, P., Khoo, J. H., Wagner, K., Liang, H., Caiazza, M. C., An, Y., Zhang, X., Chen, X., Connor-Robson, N., Shang, Z., & Wade-Martins, R. (2024). Single-cell spatial transcriptomic and translatomic profiling of dopaminergic neurons in health, aging, and disease. Cell reports, 43(3), 113784. https://doi.org/10.1016/j.celrep.2024.113784
- 发表于 2024-10-14 15:48
- 阅读 ( 1000 )
- 分类:R
你可能感兴趣的文章
- TBtools绘制火山图 11207 浏览
- 火山图--ggplot2 7327 浏览
相关问题
- 对于标准化的数据应该如何绘制火山图呢? 1 回答
0 条评论
请先 登录 后评论
