R语言绘制多分组差异分析结果图
R语言绘制多分组差异分析结果图
加载需要的包
2 设置FC和FDR的阈值线
4.3 读入数据
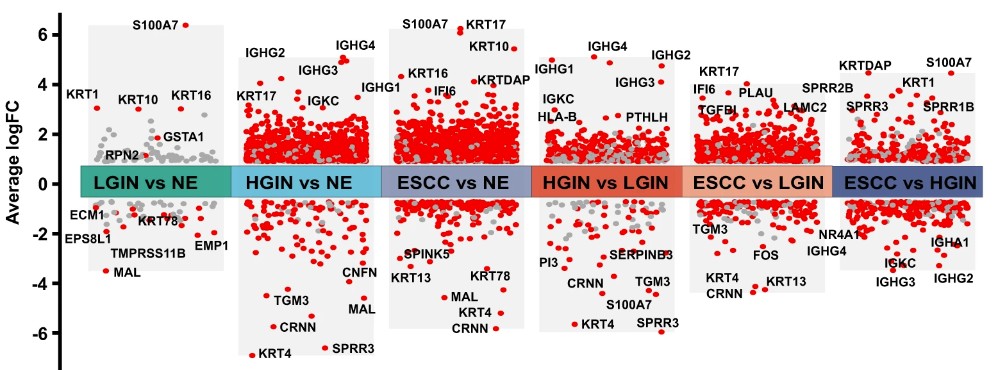
下图是一篇文献中展多分组差异分析的结果图,今天教大家如何使用R语言绘制一张类似的图。
加载需要的包
# 1.安装CRAN来源常用包
#设置镜像,
local({r <- getOption("repos")
r["CRAN"] <- "http://mirrors.tuna.tsinghua.edu.cn/CRAN/"
options(repos=r)})
# 依赖包列表:自动加载并安装
package_list <- c("ggplot2","ggnewscale","tidyverse","ggrepel","dplyr","devtools")
# 判断R包加载是否成功来决定是否安装后再加载
for(p in package_list){
if(!suppressWarnings(suppressMessages(require(p, character.only = TRUE, quietly = TRUE, warn.conflicts = FALSE)))){
install.packages(p, warn.conflicts = FALSE)
suppressWarnings(suppressMessages(library(p, character.only = TRUE, quietly = TRUE, warn.conflicts = FALSE)))
}
}
2 设置FC和FDR的阈值线
fc_cutoff <- 2 fdr_cutoff <- 0.01
3 读入数据
#设置工作路径
setwd("/share/work/wangq/piplines/class/ref/03.deg")
data<-read.table("Volcano1.txt", header=TRUE,sep = "\t")
head(data)
文件格式如下:
ID FDR log2FC cluster
Os01g0140500 1.281171e-04 -1.028091 CK_vs_KO1
Os01g0141700 9.160378e-11 1.914777 CK_vs_KO1
Os01g0339500 4.491034e-07 -2.570757 CK_vs_KO1
Os01g0392600 2.846885e-03 1.285495 CK_vs_KO1
Os01g0533900 3.601409e-07 1.708476 CK_vs_KO1
Os01g0537250 3.410239e-09 2.578599 CK_vs_KO1
4 添加上下调信息列-regulated
data <-
data %>%
mutate(change = as.factor(ifelse(FDR < fdr_cutoff & abs(log2FC) > log2(fc_cutoff),
ifelse(log2FC > log2(fc_cutoff) ,'Up','Down'),'No change'))) %>%
filter(change != 'No change') %>%
select(-change)
head(data)
data$label <- ifelse(data$FDR<0.001,"FDR<0.001","FDR>=0.001")
5 ggplot2绘图
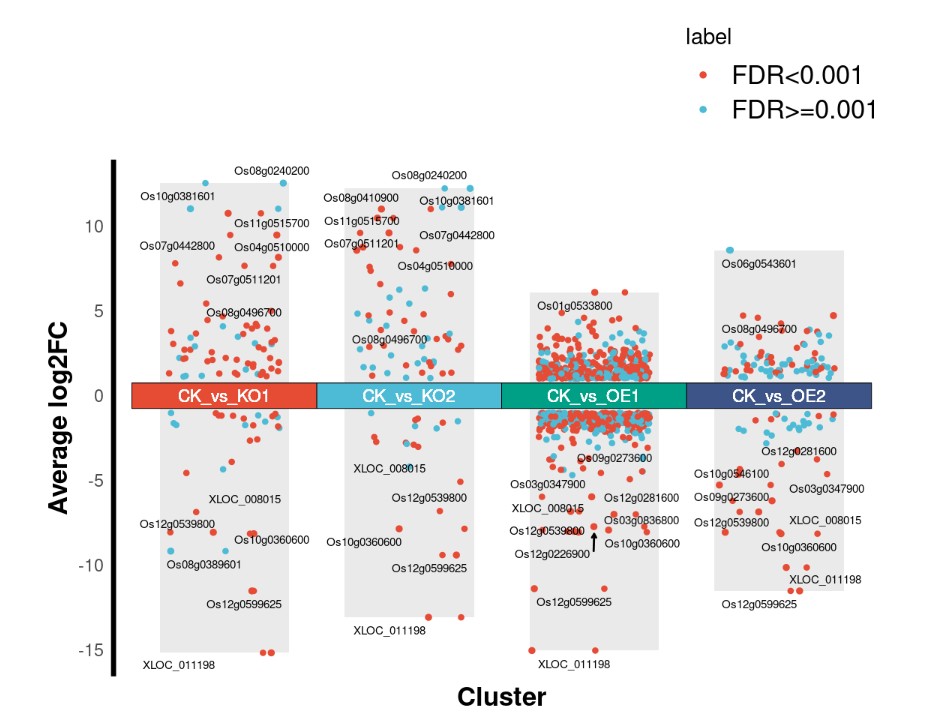
#获取每个火山图中前十个差异基因 TopGene <- data %>% group_by(cluster) %>% distinct(ID, .keep_all = T) %>% top_n(10, abs(log2FC)) table(TopGene$cluster) #背景柱状图数据准备 dbar <- data %>% group_by(cluster) %>% summarise_all(list(min = min, max = max)) %>% select(cluster, log2FC_min, log2FC_max, label_min) %>% rename(label = label_min) dbar
绘图
p <- ggplot()+
geom_col(data = dbar, # 绘制负向背景柱状图
mapping = aes(x = cluster,y = log2FC_min),
fill = "#dcdcdc",alpha = 0.6, width = 0.7) +
geom_col(data = dbar, # 绘制正向背景柱状图
mapping = aes(x = cluster,y = log2FC_max),
fill = "#dcdcdc",alpha = 0.6, width = 0.7) +
geom_jitter(data = data, # 绘制所有数据点
aes(x = cluster, y = log2FC, color = label),
size = 0.85,
width =0.3) +
geom_jitter(data = TopGene, # 绘制top10数据点
aes(x = cluster, y = log2FC, color = label),
size = 1,
width =0.35) +
geom_tile(data = TopGene, # 绘制中心分组标记图
aes(x = cluster,
y = 0,
fill = cluster),
height=1.5,
color = "black",
alpha = 0.6,
show.legend = F) +
ggsci::scale_fill_npg() + # 自定义颜色
ggsci::scale_color_npg() + # 自定义颜色
geom_text_repel(data = filter(data, ID %in% TopGene$ID), # 这里的filter很关键,筛选你想要标记的基因
aes(x = cluster, y = log2FC, label = ID),
size = 2,
max.overlaps = getOption("ggrepel.max.overlaps", default = 15),
color = 'black',
force = 1.2,
arrow = arrow(length = unit(0.008, "npc"),
type = "open", ends = "last")) +
labs(x="Cluster", y="Average log2FC") +
geom_text(data=TopGene, # 绘制中心分组标记图文本注释
aes(x=cluster,
y=0,
label=cluster),
size = 3,
color ="white") +
theme_minimal() +
theme(axis.title = element_text(size = 13,color = "black",face = "bold"),
axis.line.y = element_line(color = "black",size = 1.2),
axis.line.x = element_blank(),
axis.text.x = element_blank(),
panel.grid = element_blank(),
legend.position = "top",
legend.direction = "vertical",
legend.justification = c(1,0),
legend.text = element_text(size = 13))
p
 好了,小编就先给大家介绍到这里。希望对您的科研能有所帮助!祝您工作生活顺心快乐!
好了,小编就先给大家介绍到这里。希望对您的科研能有所帮助!祝您工作生活顺心快乐!
参考文献:Liu, X., Zhao, S., Wang, K. et al. Spatial transcriptomics analysis of esophageal squamous precancerous lesions and their progression to esophageal cancer. Nat Commun 14, 4779 (2023).
- 发表于 2024-10-25 13:52
- 阅读 ( 893 )
- 分类:R
你可能感兴趣的文章
- 跟着文献学做图 | 用蛋白质序列做主成分分析(PCA) 4786 浏览
- 使用DNA序列做主成分分析(PCA)——R语言adegenet包 5718 浏览
- R语言基础入门—矩阵介绍 3181 浏览
- R语言基础入门—数组 2503 浏览
- R语言基础入门—数据框 2057 浏览
相关问题
0 条评论
请先 登录 后评论
