安捷伦Agilent 外显子捕获试剂盒捕获区域bed文件下载
人类全外显子测序有很多测序试剂盒,比如安捷伦的SureSelect 系列试剂盒,那么他们的捕获区域可以在其官网下载:https://earray.chem.agilent.com/suredesign/index.htm
登录之后点击:"Find designs->SureSelect DNA->Agilent Catalog:
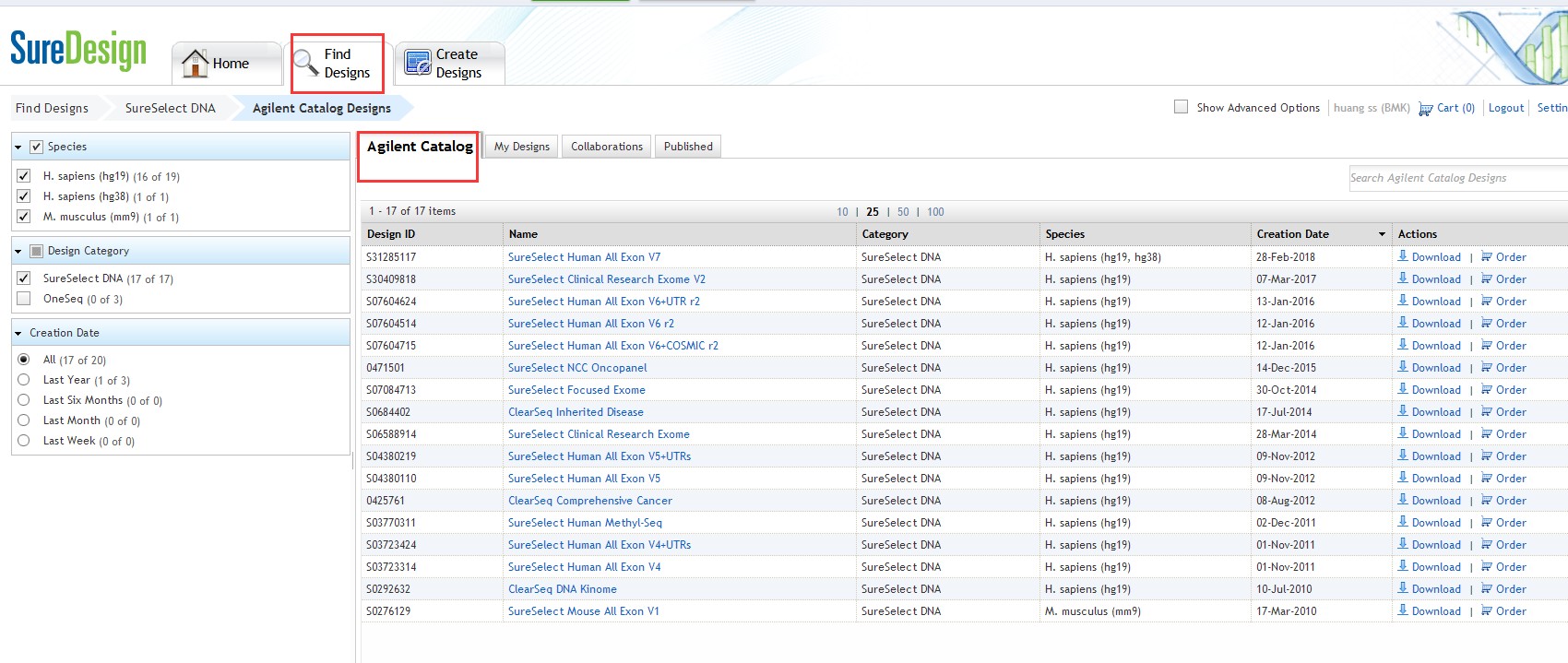 然后就得到了,各种试剂盒捕获区域文件,可以下载;当然安捷伦也支持定制panel,也就是在线自己设计panel只捕获自己想要的基因。
然后就得到了,各种试剂盒捕获区域文件,可以下载;当然安捷伦也支持定制panel,也就是在线自己设计panel只捕获自己想要的基因。
得到如下文件:
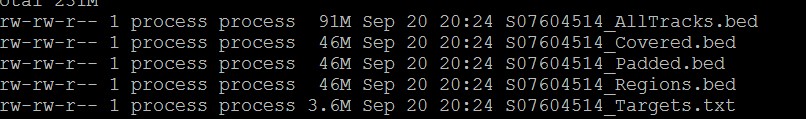
BED files
You can import these files into a compatible genome browser to graphically view the locations of the tracks in the genome.
[design ID]_Covered.bed - This BED file contains a single track of the genomic regions that are covered by one or more probes in the design. The fourth column of the file contains annotation information. You can use this file for assessing coverage metrics.
[design ID]_Padded.bed - This BED file contains a single track of the genomic regions that you can expect to sequence when using the design for target enrichment. To determine these regions, the program extends the regions in the Covered BED file by 100 bp on each side.
[design ID]_Regions.bed - This BED file contains a single track of the target regions of the design. For catalog designs, the track in this file is identical to the track in the Covered BED file.
NOTE Some SureSelect catalog designs have not been annotated to gene databases. For these designs, the gene annotation information is missing from the Covered BED file.
Text files
You can view the text files in any text editor program (e.g. NotePad) or spreadsheet program (e.g. Excel).
[design ID]_Probes.txt - This file is a list of the probes in the design, with specific information about each probe, including its probe ID, sequence, genomic coordinates, and the target it is intended to capture.
[design ID]_Targets.txt - This file contains a list of the database identifiers to which the probes in this design were annotated.
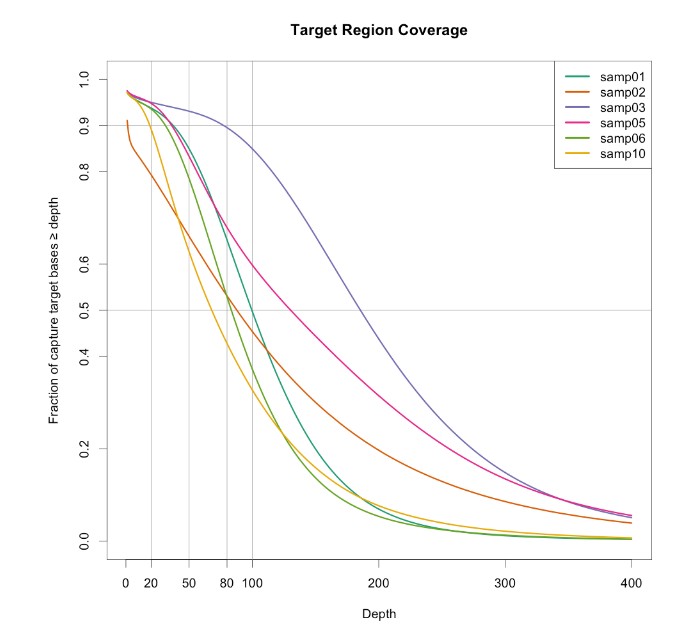
捕获区域统计
然后我们做了外显子测序,可以对捕获区域进行分析统计,工具bedtools可以实现:
参考:http://www.gettinggeneticsdone.com/2014/03/visualize-coverage-exome-targeted-ngs-bedtools.html
- 发表于 2018-09-21 10:38
- 阅读 ( 11545 )
- 分类:软件工具
