python版本MCScanX分析方法
python版本MCScanX分析方法,仅供参考:
Genomic synteny among C. annuum, C.chinenese and C.baccatum
The genomes of C. annuum, C.chinenese and C.baccatum were retrieved from PGP (http://peppergenome.snu.ac.kr/download.php).The C. annuum genome was compared to C.chinenese and C.baccatum genomes using the MCScan toolkit (V1.1)[1]. To call synteny blocks, we performed all-against-all LAST [2] and chained the LAST hits with a distance cutoff of 20 genes, also requiring at least 4 gene pairs per synteny block. Chromosome-scale synteny blocks plots were constructed using the python version of MCScan[3].
[1] Tang H, Wang X, Bowers JE, et al. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res 2008;18(12):1944–54.
[2] S. M. Kiełbasa, R. Wan, K. Sato, P. Horton, M. C. Frith, Adaptive seeds tame genomic sequence comparison. Genome Res. 21, 487–493 (2011). Medline doi:10.1101/gr.113985.110
[3] MCscan GitHub wiki https://github.com/tanghaibao/jcvi/wiki/MCscan-(Python-version) , GitHub, June 9th, 2018
参考文献:
Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome
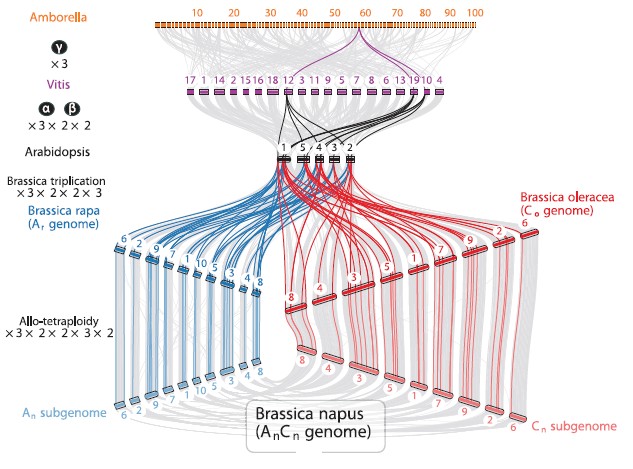
更详细的基因组共线性分析可观看:《基因家族分析实操课程》有详细操作说明
- 发表于 2018-10-16 10:01
- 阅读 ( 10839 )
- 分类:文献解读
