bam文件转换fastq
转换bam文件为fastq文件
bedtools 转换bam文件为fastq文件
bamToFastq
Tool: bedtools bamtofastq (aka bamToFastq)
Version: v2.25.0
Summary: Convert BAM alignments to FASTQ files.
Usage: bamToFastq [OPTIONS] -i <BAM> -fq <FQ>
Options:
-fq2 FASTQ for second end. Used if BAM contains paired-end data.
BAM should be sorted by query name is creating paired FASTQ.
-tags Create FASTQ based on the mate info
in the BAM R2 and Q2 tags.
Tips:
If you want to create a single, interleaved FASTQ file
for paired-end data, you can just write both to /dev/stdout:
bedtools bamtofastq -i x.bam -fq /dev/stdout -fq2 /dev/stdout > x.ilv.fq
直接转换成fastq 例如可以:
bamToFastq -i unmapped.bam -fq unmapped.fastq
结果如下
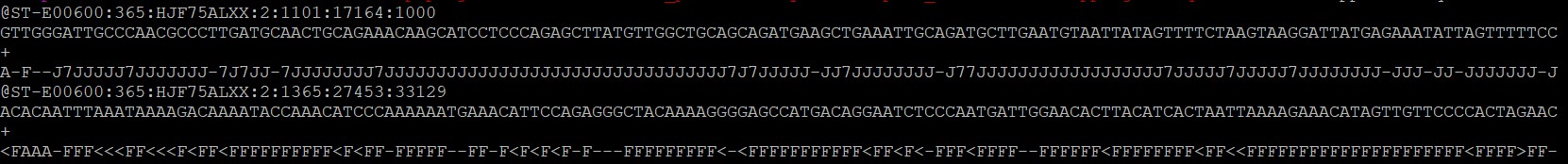
如果需要区分出bam文件中paired reads 可以:
bamToFastq -i unmapped.bam -fq unmappedRead1.fastq -fq2 unmappedRead2.fastq
- 发表于 2019-03-15 13:37
- 阅读 ( 13261 )
- 分类:软件工具
你可能感兴趣的文章
- samtools | 提取部分染色体的bam信息 1061 浏览
- 使用fastp软件对fastq文件进行剪切 1587 浏览
- bam文件格式详解 8275 浏览
- BWA比对及Samtools提取mapped reads 7403 浏览
- NCBI下载SRR并转换为fastq文件 12556 浏览
- fastq文件转换成fasta 文件perl 2279 浏览
相关问题
- 老师,在生成gbcf文件后,发现只运行了前三个染色体的,是我的代码有问题嘛?该怎么解决啊?参考基因组的fasta,fai文件都是好的,前期生成的bam文件也是好的,样品是四倍体,参考基因组是异源四倍体 1 回答
- fq.gz文件转fa文件 1 回答
- 批量srr转fastq 0 回答
- sra文件转换为fastq文件 1 回答
- fastqc运行报错是为什么?命令是:fastqc -o /root -t 2 /root/clean_reads.1.fq.gz /root/clean_reads.2.fq.gz 1 回答
- 使用bio-linux中sratoolkit里prefetch下载数据,但是无法打开试过用自带的sratoolkit,也自己去下了个最新版的放在用户文件夹也不行,我按照网上的教程设置环境变量,用source ~/.zshrc使配置生效了(好像bio-linux用bashrc不行?),但是无论如何也打不开,不知道哪里出了问题,有大佬知道吗,小白实在不懂 1 回答
0 条评论
请先 登录 后评论
