这是我的四个文件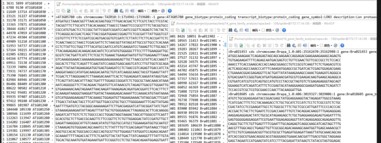
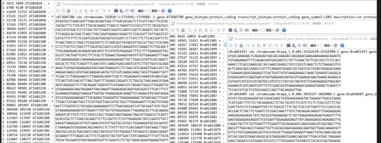
$python2 -m jcvi.compara.catalog ortholog ath rapa --cscore=0.7
12:25:58 [cbook] File `rapa.prj` exists. Computation skipped.
12:25:58 [base] lastal -u 0 -P 48 -i3G -f BlastTab rapa ath.cds >./ath.rapa.last
12:26:05 [synteny] Assuming --qbed=ath.bed --sbed=rapa.bed
12:26:05 [base] Load file `ath.bed`
12:26:05 [base] Load file `rapa.bed`
Traceback (most recent call last):
File "/home/spider/soft/miniconda3/envs/py27/lib/python2.7/runpy.py", line 174, in _run_module_as_main
"__main__", fname, loader, pkg_name)
File "/home/spider/soft/miniconda3/envs/py27/lib/python2.7/runpy.py", line 72, in _run_code
exec code in run_globals
File "/home/spider/soft/miniconda3/envs/py27/lib/python2.7/site-packages/jcvi/compara/catalog.py", line 928, in <module>
main()
File "/home/spider/soft/miniconda3/envs/py27/lib/python2.7/site-packages/jcvi/compara/catalog.py", line 80, in main
p.dispatch(globals())
File "/home/spider/soft/miniconda3/envs/py27/lib/python2.7/site-packages/jcvi/apps/base.py", line 111, in dispatch
globals[action](sys.argv[2:])
File "/home/spider/soft/miniconda3/envs/py27/lib/python2.7/site-packages/jcvi/compara/catalog.py", line 678, in ortholog
blastfilter_main([last, "--cscore={0}".format(ccscore)])
File "/home/spider/soft/miniconda3/envs/py27/lib/python2.7/site-packages/jcvi/compara/blastfilter.py", line 274, in main
blastfilter_main(blastfile, p, opts)
File "/home/spider/soft/miniconda3/envs/py27/lib/python2.7/site-packages/jcvi/compara/blastfilter.py", line 40, in blastfilter_main
qbed, sbed, qorder, sorder, is_self = check_beds(blast_file, p, opts)
File "/home/spider/soft/miniconda3/envs/py27/lib/python2.7/site-packages/jcvi/compara/synteny.py", line 490, in check_beds
sbed = Bed(opts.sbed, sorted=sorted)
File "/home/spider/soft/miniconda3/envs/py27/lib/python2.7/site-packages/jcvi/formats/bed.py", line 148, in __init__
b = BedLine(line)
File "/home/spider/soft/miniconda3/envs/py27/lib/python2.7/site-packages/jcvi/formats/bed.py", line 51, in __init__
self.start = int(args[1]) + 1
IndexError: list index out of range
看你的文件应该没有问题,再检查一下文件结尾是否有多余的空行,有的话删除试一下;
如果还不行,建议使用我们提供的biolinux 里面安装了python 的mcscan,怀疑你的软件安装问题;
https://www.omicsclass.com/article/526
基因家族分析课程:学习链接:基因家族分析实操课程、基因家族文献思路解读
如果觉得我的回答对您有用,请随意打赏。你的支持将鼓励我继续创作!
