TCGA拷贝数变异数据copy number variation 下载处理的方法不一样,你不要套用表达数据的下载方法:
TCGAbiolinks下载“Copy Number Variation”在提取矩阵时报错(在下载miRNA和甲基化数据时也遇到这个问题), expdat变量不是SummarizedExperiment这个类型,导致assay(expdat)不能提取,请问怎么解决
# 加载需要的包
library(SummarizedExperiment)
library(TCGAbiolinks)
#下载Copy Number Variation数据
query <- GDCquery(project = "TCGA-CHOL",
data.category = "Copy Number Variation",
data.type = "Copy Number Segment")
GDCdownload(query, method = "api", files.per.chunk = 50)
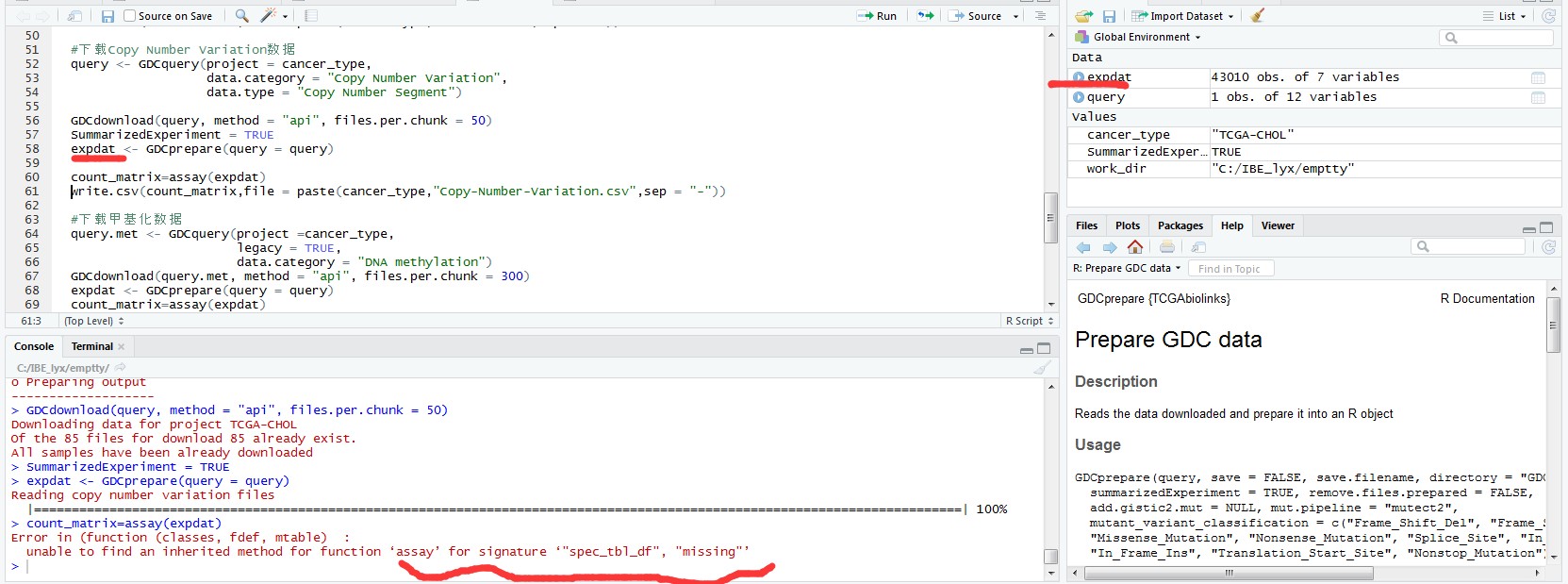
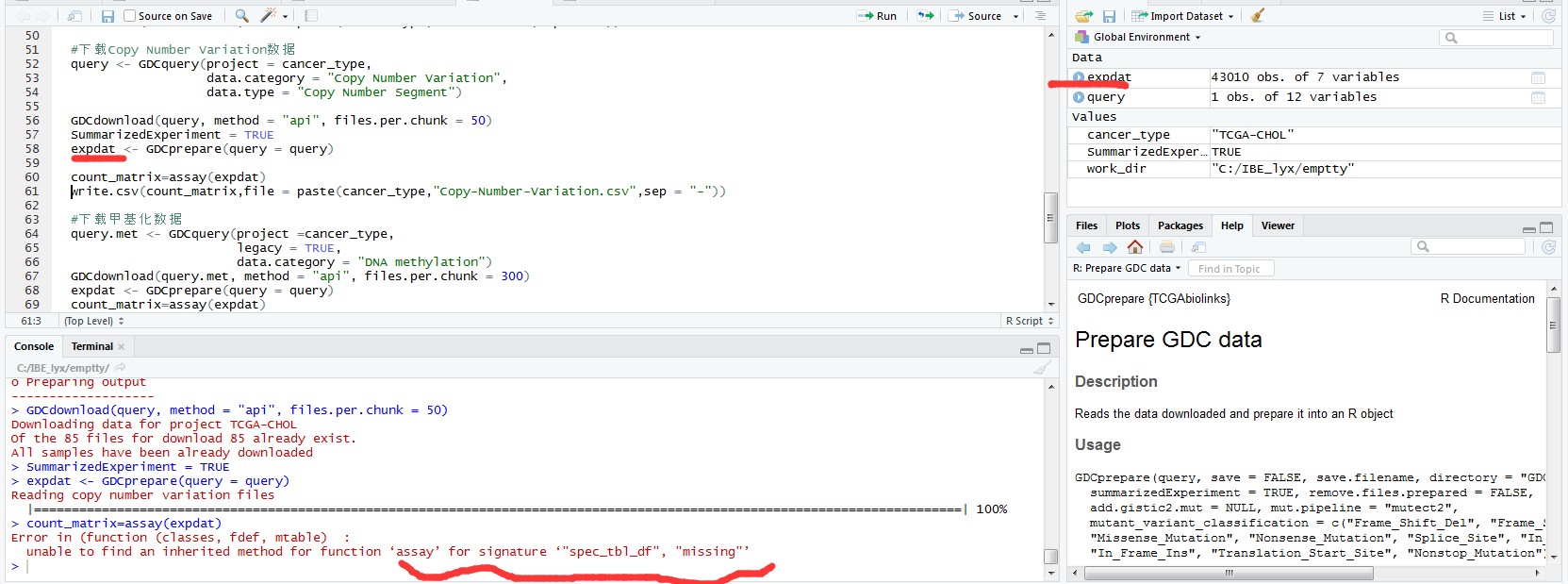
expdat <- GDCprepare(query = query)
count_matrix=assay(expdat)
write.csv(count_matrix,file = paste(cancer_type,"Copy-Number-Variation.csv",sep = "-"))
####报错
> count_matrix=assay(expdat)
Error in (function (classes, fdef, mtable) :
unable to find an inherited method for function ‘assay’ for signature ‘"spec_tbl_df", "missing"’
请先 登录 后评论
1 个回答
- 1 关注
- 0 收藏,6022 浏览
- momo 提出于 2020-03-03 00:04
相似问题
- 请教一下TCGA的蛋白数据怎么理解,它是有正负值的,这个正负值代表什么意思,为什么蛋白表达会有负值?我只想给它分成高表达,低表达和表达缺失组应该怎么处理呀? 0 回答
- TCGA下载 docker报错 0 回答
- TCGA差异分析如何根据临床信息分组? 1 回答
- TCGA用R数据下载错误 1 回答
- 老师,想请教一下TCGA基因表达数据的问题,我从xena.ucsc网页上下载了基因表达数据TCGA-CESC.htseq_counts.tsv;然后发现该数据中只有Ensembl格式的基因ID ,没有SYMBOL格式的。所以接下来进行基因ID格式转换,却发现同一个SYMBOL ID对应的多个Ensembl格式的ID,想问下老师,这种情况该怎么处理?同一个SYMBOL ID所对应的多个Ensembl格式ID的基因表达数据应该留下哪一个? 1 回答
- TCGA的RNA-seq count annotation过滤,有些不知道是否需要删除 0 回答
