这个没有测试过,你可以看看circos的官网,学习参数的设置:http://circos.ca/documentation/tutorials/
5 circos 字体大小与比例问题
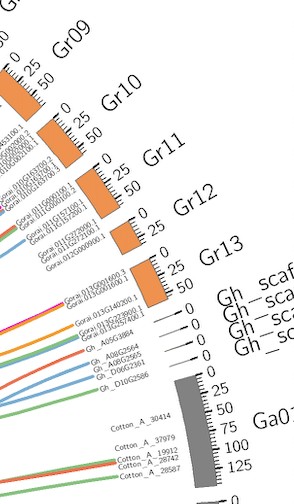
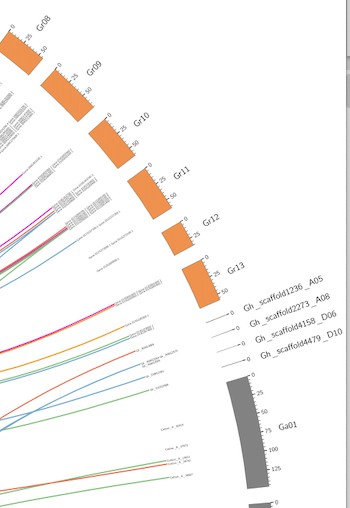 染色体的ID在图中无法展示全,我加了radius*=4000p的参数就变成第二个图,但是整体的字体缩小,看不清染色体的ID,无法一目了然,尤其是染色质刻度上。我在Circos官网上没找到,所以想问一下改哪个参数保证是第一张图的字体大小比例,还能展示全。不要像第二张图文字都包括,但字体小看不清刻度和染色体ID。
染色体的ID在图中无法展示全,我加了radius*=4000p的参数就变成第二个图,但是整体的字体缩小,看不清染色体的ID,无法一目了然,尤其是染色质刻度上。我在Circos官网上没找到,所以想问一下改哪个参数保证是第一张图的字体大小比例,还能展示全。不要像第二张图文字都包括,但字体小看不清刻度和染色体ID。
以下是第一张图的代码
chromosomes_units=1000000
chromosomes_reverse=/Ga[01]/
<ideogram>
fill=yes
label_font=default
label_parallel=no
#label_radius=dims(image,radius)-350p
label_radius=dims(image,radius)-60p
label_size=35
radius=0.90r
show_label=yes
<spacing>
default=0.005r
</spacing>
stroke_color=dgrey
stroke_thickness=2p
thickness=0.03r
</ideogram>
karyotype=/Users/823371123/Desktop/work/SBT/棉花基因家族分析/共线性分析/ghgrga/quan/chr.info.txt
<links>
bezier_radius=0r
bezier_radius_purity=0.75
color=black
crest=0.5
<link>
bezier_radius=0r
bezier_radius_purity=0.75
color=set2-8-qual-1
crest=0.5
file=/Users/823371123/Desktop/work/SBT/棉花基因家族分析/共线性分析/ghgrga/quan/SBTLickedRegion.tab.txt
radius=0.88r
#<rules>
#<rule>
#color=red
#condition=var(intrachr)
#</rule>
#<rule>
#color=red
#condition=var(interchr)
#</rule>
#</rules>
thickness=6
z=20
</link>
radius=0.40r
thickness=1
</links>
<plots>
<plot>
#color=set2-8-qual-2
color=black
file=/Users/823371123/Desktop/work/SBT/棉花基因家族分析/共线性分析/ghgrga/quan/textSBT.txt
label_font=light
link_color=black
link_dims=0p,2p,5p,2p,2p
link_thickness=2p
label_snuggle=yes
r0=0.88r
r1=0.99r
rpadding=5p
show_links=no
type=text
</plot>
type=histogram
</plots>
show_tick_labels=yes
show_ticks=yes
spacing=10u
<ticks>
color=black
format=%d
multiplier=1e-6
radius=1r
thickness=2p
<tick>
size=10p
spacing=5u
</tick>
<tick>
color=black
format=%d
label_offset=10p
label_size=25p
show_label=yes
size=15p
spacing=25u
thickness=4p
</tick>
</ticks>
<colors>
<<include etc/colors.conf>>
<<include etc/brewer.conf>>
#<<include etc/colors_fonts_patterns.conf>>
#<<include colors.ucsc.conf>>
#<<include colors.hsv.conf>>
</colors>
<fonts>
<<include etc/fonts.conf>>
</fonts>
<image>
<<include etc/image.conf>>
#radius*=4000p
</image>
<<include etc/housekeeping.conf>>
1 个回答
- 1 关注
- 0 收藏,2917 浏览
- [玺] 提出于 2021-11-25 13:36
