刚刚测试了我们的镜像,没问题的,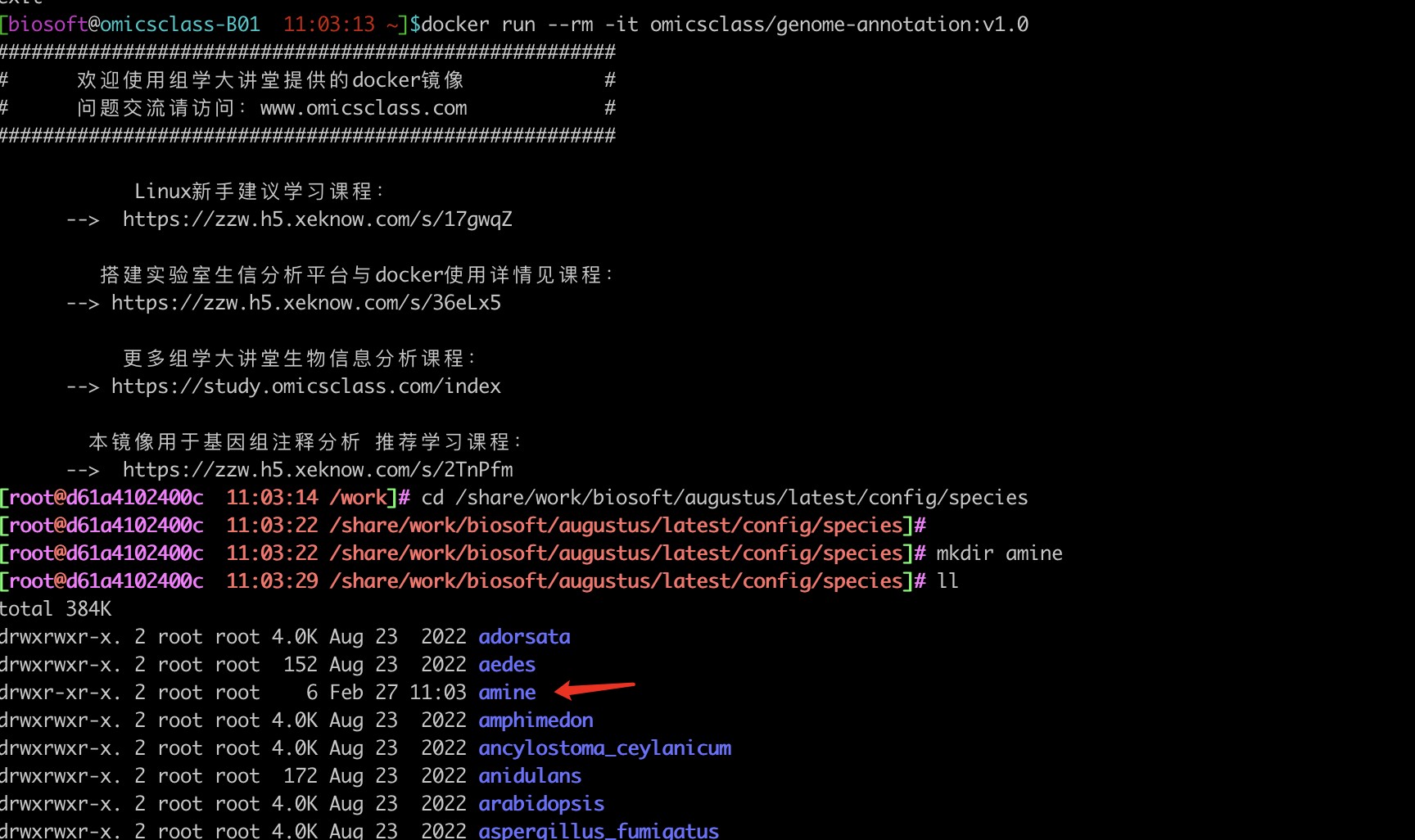
动植物基因组组装课程,结构注释部分,运行braker.pl出错
脚本:03.Braker.sh 第78行:
braker.pl --species=$species \
--genome=$contig \
--softmasking \
--bam=rnaseq.bam \
--cores=$threads \
--useexisting \
--gff3 --skip_fixing_broken_genes
系统提示:
Failed to create new species with new_species.pl, check write permissions in /share/work/biosoft/augustus/latest/config//species directory! Command was perl /share/work/biosoft/augustus/augustus-3.3.3/scripts/new_species.pl --species=Acer_truncatum --AUGUSTUS_CONFIG_PATH=/share/work/biosoft/augustus/latest/config/ 1> /dev/null 2>/home/shpc_100675/workspace/zxdjt/animal/annotation/03.Braker/braker/errors/new_species.stderr
问题:
1、/share/work/biosoft/augustus/latest/config//species :contig后面怎么有两个斜线?用cd命令,正常应该是/share/work/biosoft/augustus/latest/config/species
2、根据提示,查看了目录species的权限,发现是可读可写可执行属性(drwxrwxrwx 111 root root 2556 Aug 23 2022 species),但是在species内新建目录,提示如下。怎么解决不能写入文件的问题?
mkdir mine
mkdir: cannot create directory ‘mine’: Read-only file system
1 个回答
- 1 关注
- 0 收藏,2314 浏览
- cashing 提出于 2023-02-27 00:15
