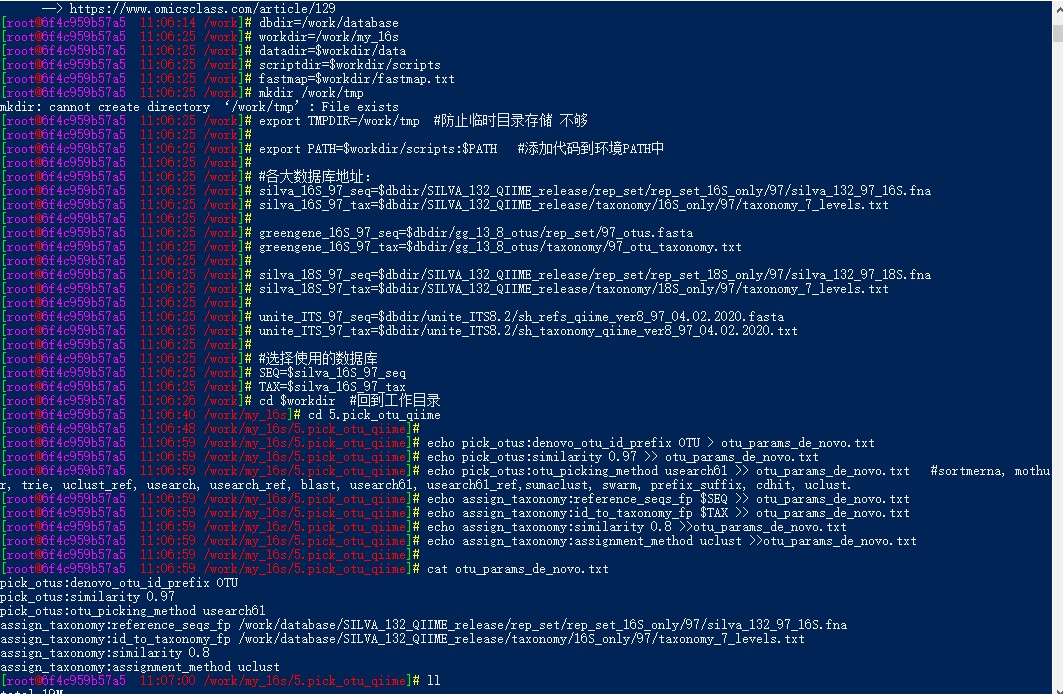
100 docker运行qiime1时按照教程进行到章节2课时6时,即聚类生成OTU-denovo时老是报错,查看类似问题后,升级了运行内存为70g,同时将教程内的30多个样本减成2个样本,FASTQ格式还有qimme格式检查都正确,依旧报错,报错内容如下:Segmentation fault
docker运行qiime1时按照教程进行到章节2课时6时,即聚类生成OTU-denovo时老是报错,查看类似问题后,升级了运行内存为70g,同时将教程内的30多个样本减成2个样本,FASTQ格式还有qimme格式检查都正确,依旧报错,报错内容如下:
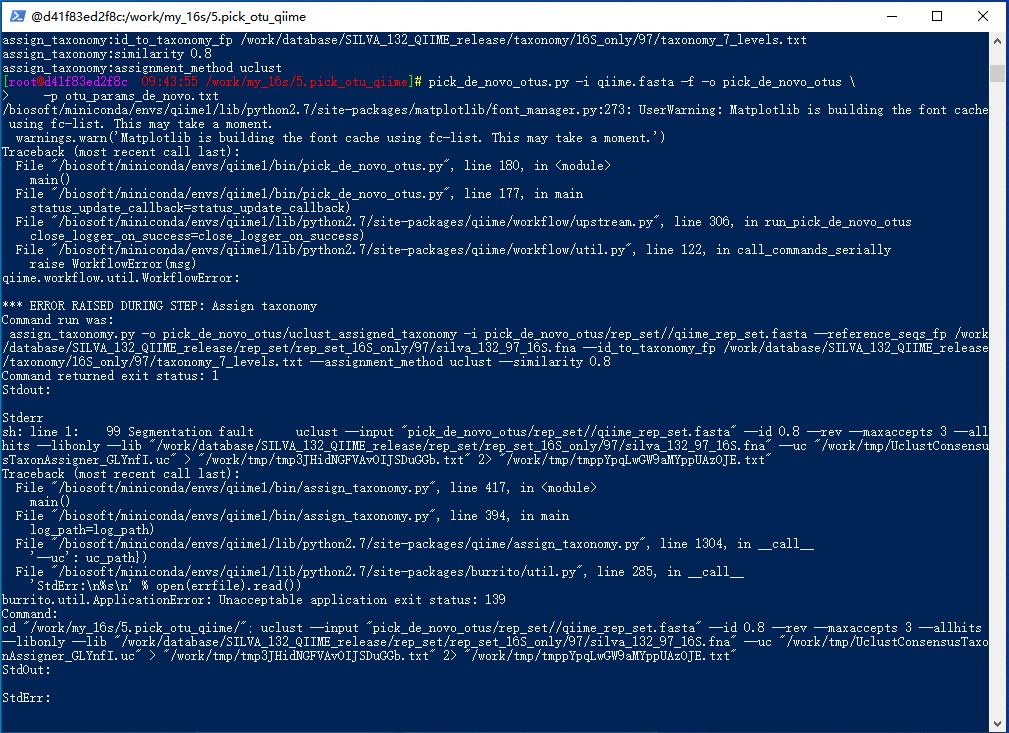
pick_de_novo_otus.py -i qiime.fasta -f -o pick_de_novo_otus \
> -p otu_params_de_novo.txt
/biosoft/miniconda/envs/qiime1/lib/python2.7/site-packages/matplotlib/font_manager.py:273: UserWarning: Matplotlib is building the font cache using fc-list. This may take a moment.
warnings.warn('Matplotlib is building the font cache using fc-list. This may take a moment.')
Traceback (most recent call last):
File "/biosoft/miniconda/envs/qiime1/bin/pick_de_novo_otus.py", line 180, in <module>
main()
File "/biosoft/miniconda/envs/qiime1/bin/pick_de_novo_otus.py", line 177, in main
status_update_callback=status_update_callback)
File "/biosoft/miniconda/envs/qiime1/lib/python2.7/site-packages/qiime/workflow/upstream.py", line 306, in run_pick_de_novo_otus
close_logger_on_success=close_logger_on_success)
File "/biosoft/miniconda/envs/qiime1/lib/python2.7/site-packages/qiime/workflow/util.py", line 122, in call_commands_serially
raise WorkflowError(msg)
qiime.workflow.util.WorkflowError:
*** ERROR RAISED DURING STEP: Assign taxonomy
Command run was:
assign_taxonomy.py -o pick_de_novo_otus/uclust_assigned_taxonomy -i pick_de_novo_otus/rep_set//qiime_rep_set.fasta --reference_seqs_fp /work/database/SILVA_132_QIIME_release/rep_set/rep_set_16S_only/97/silva_132_97_16S.fna --id_to_taxonomy_fp /work/database/SILVA_132_QIIME_release/taxonomy/16S_only/97/taxonomy_7_levels.txt --assignment_method uclust --similarity 0.8
Command returned exit status: 1
Stdout:
Stderr
sh: line 1: 98 Segmentation fault uclust --input "pick_de_novo_otus/rep_set//qiime_rep_set.fasta" --id 0.8 --rev --maxaccepts 3 --allhits --libonly --lib "/work/database/SILVA_132_QIIME_release/rep_set/rep_set_16S_only/97/silva_132_97_16S.fna" --uc "/work/tmp/UclustConsensusTaxonAssigner_jC5NV4.uc" > "/work/tmp/tmpsnWnyn7pct7xAzj5h5pm.txt" 2> "/work/tmp/tmpHGfUbEEXt29bAiXMuOoV.txt"
Traceback (most recent call last):
File "/biosoft/miniconda/envs/qiime1/bin/assign_taxonomy.py", line 417, in <module>
main()
File "/biosoft/miniconda/envs/qiime1/bin/assign_taxonomy.py", line 394, in main
log_path=log_path)
File "/biosoft/miniconda/envs/qiime1/lib/python2.7/site-packages/qiime/assign_taxonomy.py", line 1304, in __call__
'--uc': uc_path})
File "/biosoft/miniconda/envs/qiime1/lib/python2.7/site-packages/burrito/util.py", line 285, in __call__
'StdErr:\n%s\n' % open(errfile).read())
burrito.util.ApplicationError: Unacceptable application exit status: 139
Command:
cd "/work/my_16s/5.pick_otu_qiime/"; uclust --input "pick_de_novo_otus/rep_set//qiime_rep_set.fasta" --id 0.8 --rev --maxaccepts 3 --allhits --libonly --lib "/work/database/SILVA_132_QIIME_release/rep_set/rep_set_16S_only/97/silva_132_97_16S.fna" --uc "/work/tmp/UclustConsensusTaxonAssigner_jC5NV4.uc" > "/work/tmp/tmpsnWnyn7pct7xAzj5h5pm.txt" 2> "/work/tmp/tmpHGfUbEEXt29bAiXMuOoV.txt"
StdOut:
StdErr:
- 1 关注
- 1 收藏,2867 浏览
- Lucian 提出于 2023-06-04 11:20
可能是软件编译问题,你换个电脑试试
用了两台电脑都是这样的报错,最后用linux系统重新运行,就可以了,windows系统还是容易出现这些乱七八糟的问题