最好有截图说明自己的问题。
下面的配置文件可以绘制如下图片,你可以试一下:
更多见:《基因家族分析实操课程》:
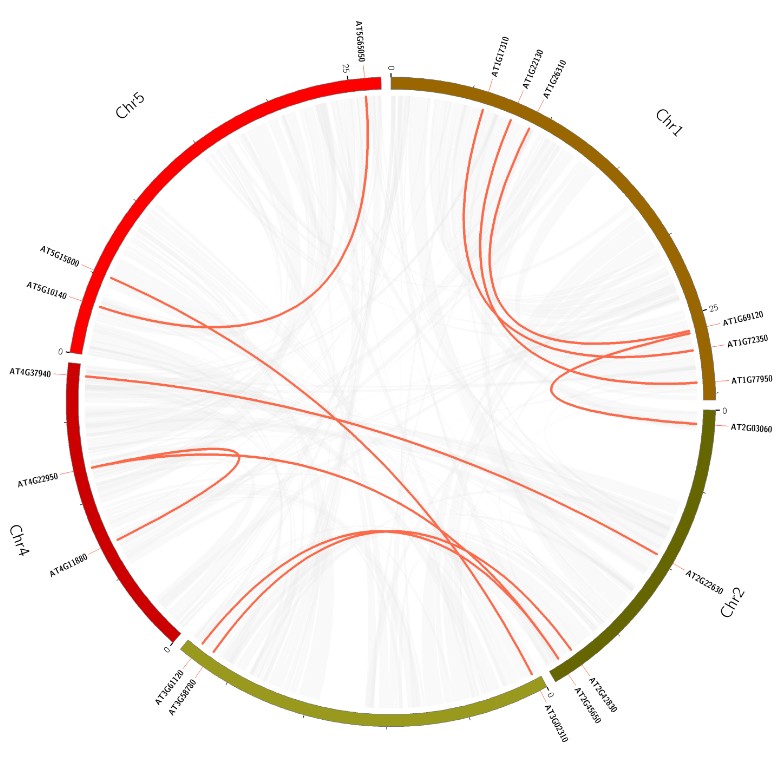
chromosomes_units=1000000 #刻度单位Mb
#chromosomes_reverse=/[12345]/ #染色体反转
karyotype=./chr.info #染色体信息配置文件
show_tick_labels=yes
show_ticks=yes
spacing=10u
<ticks> #设置染色体刻度
color=black
format=%d
multiplier=1e-6
radius=1r
thickness=2p
<tick>
size=10p
spacing=5u
</tick>
<tick>
color=black
format=%d
label_offset=10p
label_size=25p
show_label=yes
size=15p
spacing=25u
thickness=4p
</tick>
</ticks>
<ideogram> #染色体绘制设置
fill=yes #是否填充颜色
label_font=default
label_parallel=yes
label_radius=dims(image,radius)-60p
label_size=45
radius=0.8r #设置半径,以免基因名称过长超出显示范围
show_label=yes
<spacing>
default=0.005r
</spacing>
stroke_color=dgrey
stroke_thickness=2p
thickness=0.03r
</ideogram>
<links> #设置连线
bezier_radius=0r
bezier_radius_purity=0.75
color=black
crest=0.5
<link> #基因家族共线性
bezier_radius=0r
bezier_radius_purity=0.75
color=set2-8-qual-1
crest=0.5
file=./data/MADS.link.txt
radius=0.98r
<rules>
<rule>
color=red
condition=var(intrachr)
</rule>
<rule>
color=red
condition=var(interchr)
</rule>
</rules>
thickness=8
z=20 #层数
</link>
<link> #全基因组共线性
bezier_radius=0r
bezier_radius_purity=0.75
color=230,230,230,0.2
crest=0.5
ribbon=yes
file=./data/genome.blocklink.txt
radius=0.98r
thickness=1
z=15
</link>
radius=0.40r
thickness=1
</links>
<plots>
<plot>
color=black
file=./data/MADS.text.txt
label_font=condensed
label_size=24p
link_color=red
link_dims=0p,0p,50p,0p,10p
link_thickness=2p
r0=1r
r1=1r+200p
rpadding=0p
padding=0p
show_links=yes
type=text
</plot>
type=histogram
</plots>
<colors>
<<include etc/colors.conf>>
<<include etc/brewer.conf>>
#<<include etc/colors_fonts_patterns.conf>>
#<<include colors.ucsc.conf>>
#<<include colors.hsv.conf>>
</colors>
<fonts>
<<include etc/fonts.conf>>
</fonts>
<image>
<<include etc/image.conf>>
</image>
<<include etc/housekeeping.conf>>
