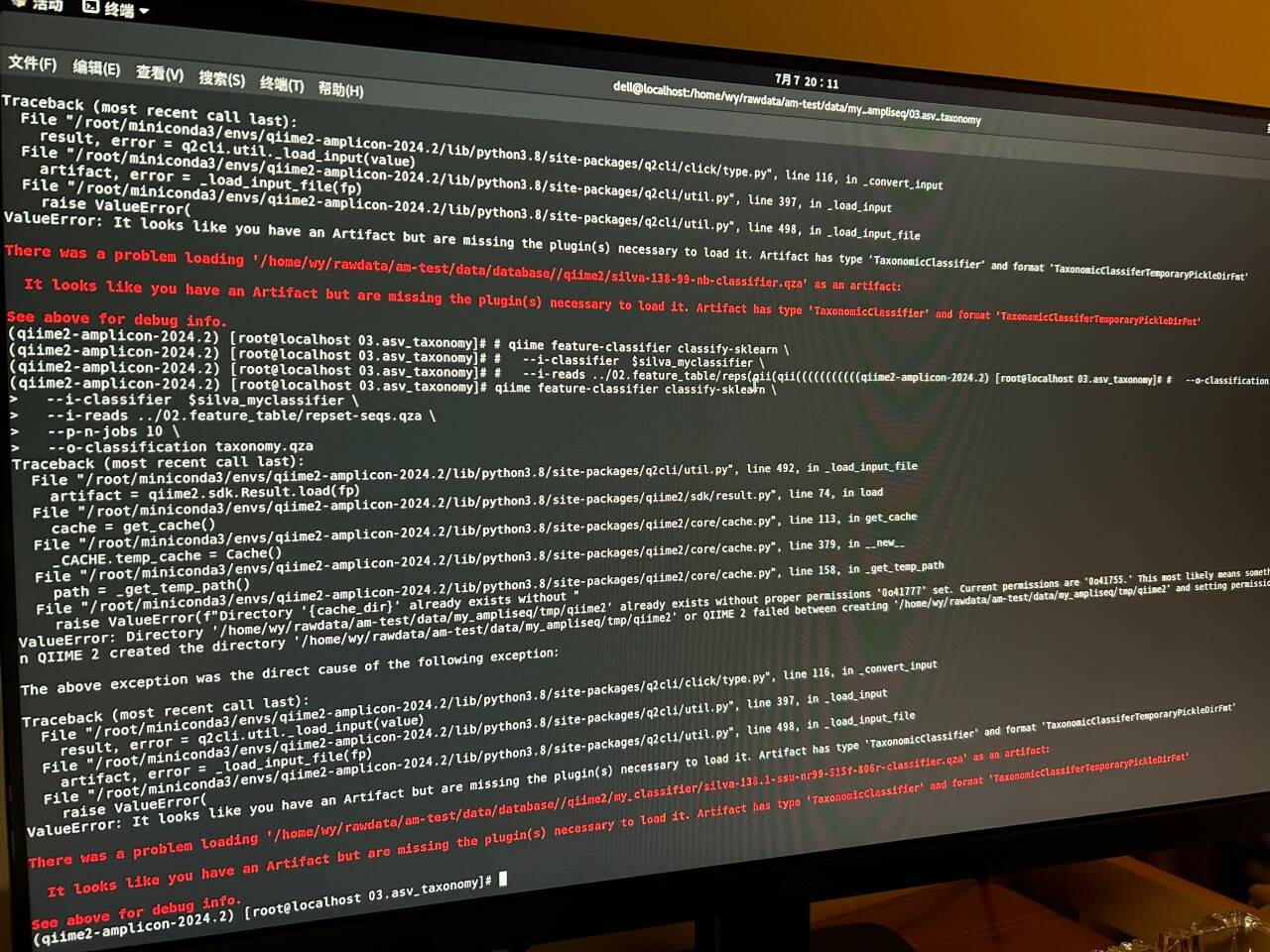
asv分类报错
输入:
qiime feature-classifier classify-sklearn \
--i-classifier $silva_classifier \
--i-reads ../02.feature_table/repset-seqs.qza \
--p-n-jobs 10 \
--p-confidence 0.7 \
--o-classification taxonomy.qza
报错
Traceback (most recent call last):
File "/root/miniconda3/envs/qiime2-amplicon-2024.2/lib/python3.8/site-packages/q2cli/util.py", line 492, in _load_input_file
artifact = qiime2.sdk.Result.load(fp)
File "/root/miniconda3/envs/qiime2-amplicon-2024.2/lib/python3.8/site-packages/qiime2/sdk/result.py", line 74, in load
cache = get_cache()
File "/root/miniconda3/envs/qiime2-amplicon-2024.2/lib/python3.8/site-packages/qiime2/core/cache.py", line 113, in get_cache
_CACHE.temp_cache = Cache()
File "/root/miniconda3/envs/qiime2-amplicon-2024.2/lib/python3.8/site-packages/qiime2/core/cache.py", line 379, in __new__
path = _get_temp_path()
File "/root/miniconda3/envs/qiime2-amplicon-2024.2/lib/python3.8/site-packages/qiime2/core/cache.py", line 158, in _get_temp_path
raise ValueError(f"Directory '{cache_dir}' already exists without "
ValueError: Directory '/home/wy/rawdata/am-test/data/my_ampliseq/tmp/qiime2' already exists without proper permissions '0o41777' set. Current permissions are '0o41755.' This most likely means something other than QIIME 2 created the directory '/home/wy/rawdata/am-test/data/my_ampliseq/tmp/qiime2' or QIIME 2 failed between creating '/home/wy/rawdata/am-test/data/my_ampliseq/tmp/qiime2' and setting permissions on it.
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/root/miniconda3/envs/qiime2-amplicon-2024.2/lib/python3.8/site-packages/q2cli/click/type.py", line 116, in _convert_input
result, error = q2cli.util._load_input(value)
File "/root/miniconda3/envs/qiime2-amplicon-2024.2/lib/python3.8/site-packages/q2cli/util.py", line 397, in _load_input
artifact, error = _load_input_file(fp)
File "/root/miniconda3/envs/qiime2-amplicon-2024.2/lib/python3.8/site-packages/q2cli/util.py", line 498, in _load_input_file
raise ValueError(
ValueError: It looks like you have an Artifact but are missing the plugin(s) necessary to load it. Artifact has type 'TaxonomicClassifier' and format 'TaxonomicClassiferTemporaryPickleDirFmt'
There was a problem loading '/home/wy/rawdata/am-test/data/database//qiime2/silva-138-99-nb-classifier.qza' as an artifact:
It looks like you have an Artifact but are missing the plugin(s) necessary to load it. Artifact has type 'TaxonomicClassifier' and format 'TaxonomicClassiferTemporaryPickleDirFmt'
See above for debug info.
2 个回答
- 2 关注
- 0 收藏,1634 浏览
- wyi0114 提出于 2024-07-08 10:46
