重复序列分析,去除含有基因序列的库文件时候报错,你们示例数据也有这样的问题
基因组重复序列分析的时候,在运行【汇总 不同软件生成的repeat库,并用RepeatMasker进行重复序列注释】时候
for lib in ModelerAll.lib MITE_LTR.lib Homology.db; do
blastx -query ${lib} -db ${SPROT} -evalue 1e-10 -num_descriptions 10 -num_threads ${threads} -out ${lib}_blast_results.txt
perl $scriptsdir/ProtExcluder1.2/ProtExcluder.pl ${lib}_blast_results.txt ${lib}
echo -e "${lib}\tbefore\t$(grep -c ">" ${lib})\tafter\t$(grep -c ">" ${lib}noProtFinal)"
done
会报错如下:
Can not open the seqfile ModelerAll.lib_blast_results.txt.fnolowm50seq
mergeunmatchedregion.pl seqfile
Illegal division by zero at /public-supool/home/thli/scripts/genome_annotation/ProtExcluder1.2/GCcontent.pl line 122.
Can not open the seqfile MITE_LTR.lib_blast_results.txt.fnolowm50seq
mergeunmatchedregion.pl seqfile
Illegal division by zero at /public-supool/home/thli/scripts/genome_annotation/ProtExcluder1.2/GCcontent.pl line 122.
Can not open the seqfile Homology.db_blast_results.txt.fnolowm50seq
mergeunmatchedregion.pl seqfile
Illegal division by zero at /public-supool/home/thli/scripts/genome_annotation/ProtExcluder1.2/GCcontent.pl line 122.
vsearch v2.18.0_linux_x86_64, 503.4GB RAM, 128 cores
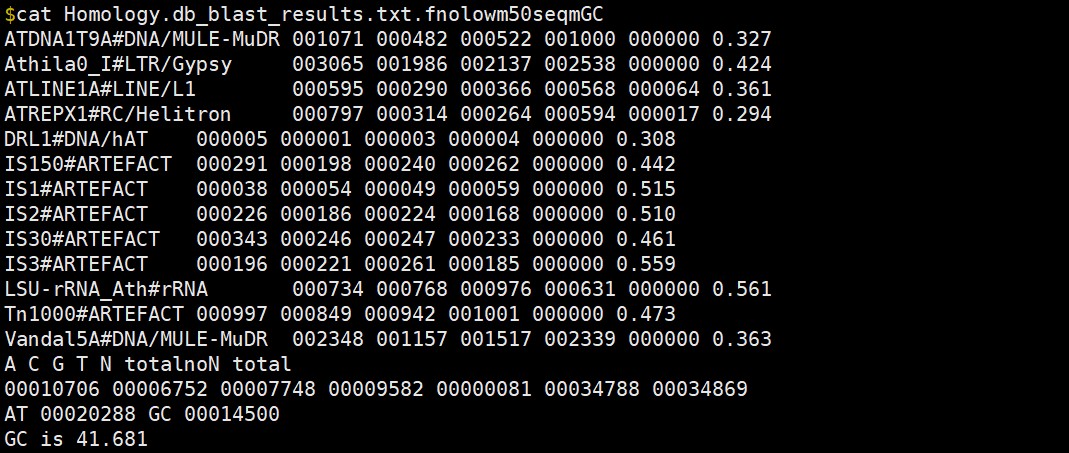
我看了你们示例输出文件中的Homology.db_blast_results.txt.fnolowm50seqmGC,发现输出都是:
A C G T N totalnoN total
00000000 00000000 00000000 00000000 00000000 00000000 00000000
AT 00000000 GC 00000000
明显你们的示例数据也计算错误。是不是脚本有问题
请先 登录 后评论
2 个回答
*_blast_results.txt输出如下:
cmd>head -50 Homology.db_blast_results.txt
BLASTX 2.14.1+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Database: uniprot_sprot_clean.fasta
565,168 sequences; 203,477,143 total letters
Query= IS1#ARTEFACT @root [S:10]
Length=768
Score E
Sequences producing significant alignments: (Bits) Value
sp|P59843|INSB_HAEDU 348 2e-122
sp|A0A385XJL4|INSB9_ECOLI 348 2e-122
sp|P0CF30|INSB8_ECOLI 348 2e-122
sp|P0CF29|INSB6_ECOLI 348 2e-122
sp|P0CF28|INSB5_ECOLI 348 2e-122
sp|P0CF25|INSB1_ECOLI 348 2e-122
sp|P0CF31|INSB_ECOLX 346 2e-121
sp|P57998|INSB4_ECOLI 338 3e-118
sp|P0CF27|INSB3_ECOLI 335 3e-117
sp|P0CF26|INSB2_ECOLI 335 3e-117
>sp|P59843|INSB_HAEDU
Length=167
Score = 348 bits (893), Expect = 2e-122, Method: Compositional matrix adjust.
Identities = 167/167 (100%), Positives = 167/167 (100%), Gaps = 0/167 (0%)
Frame = +1
Query 250 MPGNSPHYGRWPQHDFTSLKKLRPQSVTSRIQPGSDVIVCAEMDEQWGYVGAKSRQRWLF 429
MPGNSPHYGRWPQHDFTSLKKLRPQSVTSRIQPGSDVIVCAEMDEQWGYVGAKSRQRWLF
Sbjct 1 MPGNSPHYGRWPQHDFTSLKKLRPQSVTSRIQPGSDVIVCAEMDEQWGYVGAKSRQRWLF 60
Query 430 YAYDSLRKTVVAHVFGERTMATLGRLMSLLSPFDVVIWMTDGWPLYESRLKGKLHVISKR 609
YAYDSLRKTVVAHVFGERTMATLGRLMSLLSPFDVVIWMTDGWPLYESRLKGKLHVISKR
Sbjct 61 YAYDSLRKTVVAHVFGERTMATLGRLMSLLSPFDVVIWMTDGWPLYESRLKGKLHVISKR 120
Query 610 YTQRIERHNLNLRQHLARLGRKSLSFSKSVELHDKVIGHYLNIKHYQ 750
YTQRIERHNLNLRQHLARLGRKSLSFSKSVELHDKVIGHYLNIKHYQ
*_blast_results.txt输出正常,但是Homology.db_blast_results.txt.fnolowm50seqmGC,输出还是错误:
A C G T N totalnoN total
00000000 00000000 00000000 00000000 00000000 00000000 00000000
AT 00000000 GC 00000000
请先 登录 后评论
- 1 关注
- 0 收藏,940 浏览
- litianhuan 提出于 2024-09-10 22:29
*_blast_results.txt输出如下: cmd>head -50 Homology.db_blast_results.txt BLASTX 2.14.1+ Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A. Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Database: uniprot_sprot_clean.fasta 565,168 sequences; 203,477,143 total letters Query= IS1#ARTEFACT @root [S:10] Length=768 Score E Sequences producing significant alignments: (Bits) Value sp|P59843|INSB_HAEDU 348 2e-122 sp|A0A385XJL4|INSB9_ECOLI 348 2e-122 sp|P0CF30|INSB8_ECOLI 348 2e-122 sp|P0CF29|INSB6_ECOLI 348 2e-122 sp|P0CF28|INSB5_ECOLI 348 2e-122 sp|P0CF25|INSB1_ECOLI 348 2e-122 sp|P0CF31|INSB_ECOLX 346 2e-121 sp|P57998|INSB4_ECOLI 338 3e-118 sp|P0CF27|INSB3_ECOLI 335 3e-117 sp|P0CF26|INSB2_ECOLI 335 3e-117 >sp|P59843|INSB_HAEDU Length=167 Score = 348 bits (893), Expect = 2e-122, Method: Compositional matrix adjust. Identities = 167/167 (100%), Positives = 167/167 (100%), Gaps = 0/167 (0%) Frame = +1 Query 250 MPGNSPHYGRWPQHDFTSLKKLRPQSVTSRIQPGSDVIVCAEMDEQWGYVGAKSRQRWLF 429 MPGNSPHYGRWPQHDFTSLKKLRPQSVTSRIQPGSDVIVCAEMDEQWGYVGAKSRQRWLF Sbjct 1 MPGNSPHYGRWPQHDFTSLKKLRPQSVTSRIQPGSDVIVCAEMDEQWGYVGAKSRQRWLF 60 Query 430 YAYDSLRKTVVAHVFGERTMATLGRLMSLLSPFDVVIWMTDGWPLYESRLKGKLHVISKR 609 YAYDSLRKTVVAHVFGERTMATLGRLMSLLSPFDVVIWMTDGWPLYESRLKGKLHVISKR Sbjct 61 YAYDSLRKTVVAHVFGERTMATLGRLMSLLSPFDVVIWMTDGWPLYESRLKGKLHVISKR 120 Query 610 YTQRIERHNLNLRQHLARLGRKSLSFSKSVELHDKVIGHYLNIKHYQ 750 YTQRIERHNLNLRQHLARLGRKSLSFSKSVELHDKVIGHYLNIKHYQ *_blast_results.txt是否正常,Homology.db_blast_results.txt.fnolowm50seqmGC,输出还是错误: A C G T N totalnoN total 00000000 00000000 00000000 00000000 00000000 00000000 00000000 AT 00000000 GC 00000000
手动运行一下perl $scriptsdir/ProtExcluder1.2/ProtExcluder.pl MITE_LTR.lib_blast_results.txt MITE_LTR.lib,我刚刚在docker里面运行这一行是可以正常出来结果的。看你的报错不在docker的环境里,在运行过程中可能由于缺少依赖和环境变量导致某一步未能正常运算