150 GO注释:blast了nr数据库并通过blast2go获得了GO注释号;也通过blast了swiss-prot数据库获得了GO注释号。这两个结果需要合并吗?
老师,你好,
首先,我在本地将我的序列blast了nr数据库,并导入blast2go软件进行了GO注释,并获得了go注释号。另外,我还将数据在本地比对swiss-prot数据库并进一步获得了GO注释号。
1. 我想在想讲这个结果导入WEGO进行作图,请问上面两份结果需要怎么处理,是需要合并吗,怎么合并?
我是参考一片文献进行操作的,他的描述如下:CDS sequences with SSR were aligned against NCBI non-redundant (nr) and SWISS-PROT protein databases (http://www.uniprot.org), using BLASTx (Altschul et al., 1997) with a cutoff E-value 1E-5. Blast results were imported into Blast2Go (Conesa et al., 2005) for GO term mapping. Mapping results were submitted to WEGO (Ye et al., 2006) for GO classification.
他的结果如下:
814 and 745 CDSs with SSR in genomes of C. exilicauda and M. martensii, respectively, were subjected to GO analysis based on sequence alignment and the results were used in GO enrichment analysis. In total, 261 (C. exilicauda) and 197 (M. martensii) CDSs were assigned with 1562 (C. exilicauda) and 1390 (M. martensii) GO terms of known
function. The percentages of CDSs with SSR assigned to each subcategory are shown in Fig. 4. Comparison of function distribution between these two species demonstrated that only CDSs of C. exilicauda were assigned to auxiliary transport protein (GO: 0015457) and rhythmic process (GO: 0048511). Only CDSs of M. martensii were assigned to virion (GO: 0,019,012).
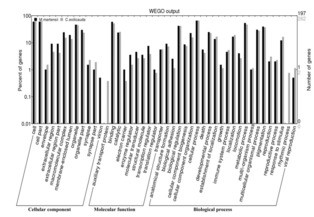
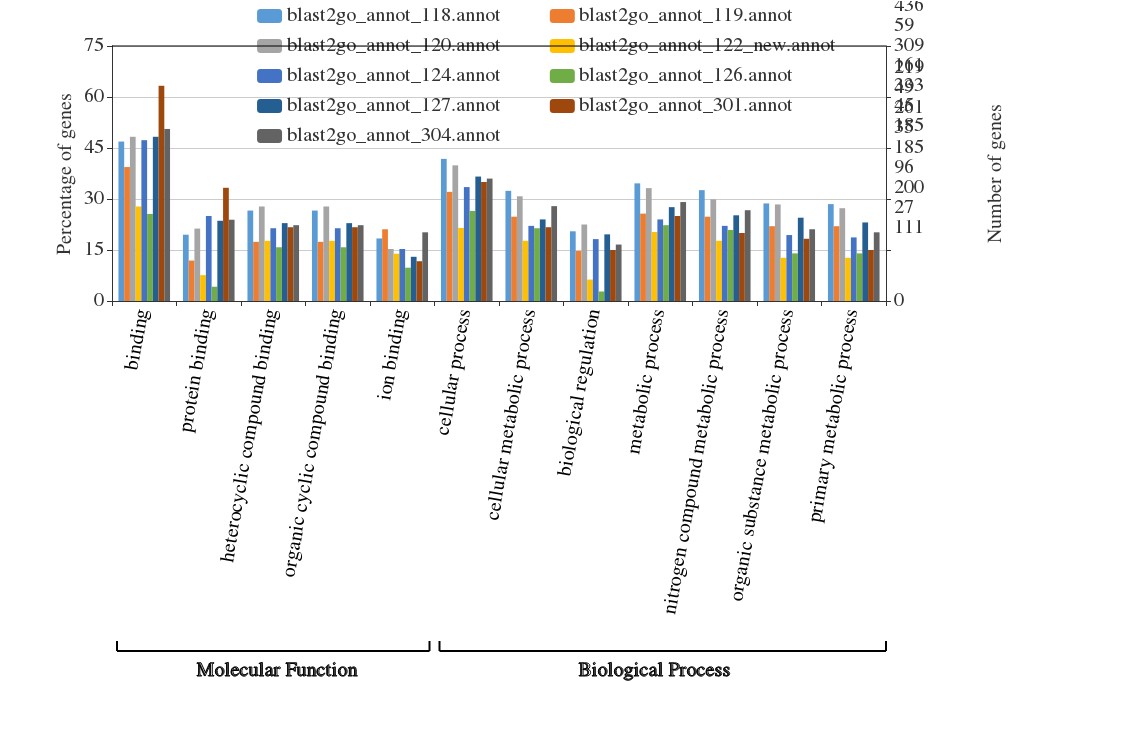
2. 但奇怪的是:我把我从blast了nr数据库获得的GO注释号的几个文件导入WEGO后,发现缺少细胞组分这一块(如果我只放单个文件进去就会有CP,MF和BP),这又是什么问题?
- 2 关注
- 0 收藏,7698 浏览
- Mayomics 提出于 2019-05-29 21:40
相似问题
- 转录组差异分析里GO注释的图怎么看的? 1 回答
- blast2go注释,报错“Exception in thread "main" java.lang.NoClassDefFoundError: org/jdom/JDOMException”,请问如何解决,详细一点 1 回答
- Blast2go本地化过程中,运行官方文件b2g4pipe_v2.5.zip中“runPipeExample.sh“报错“The host did not accept the connection within timeout of 60000 ms;” 1 回答
- blast2go本地化过程中,导入数据库不全,请问老师是哪一个数据库出了问题?我还想要一份无参GO注释的流程或者手册之类的,谢谢 1 回答
- 如何进行go的注释和富集 1 回答
