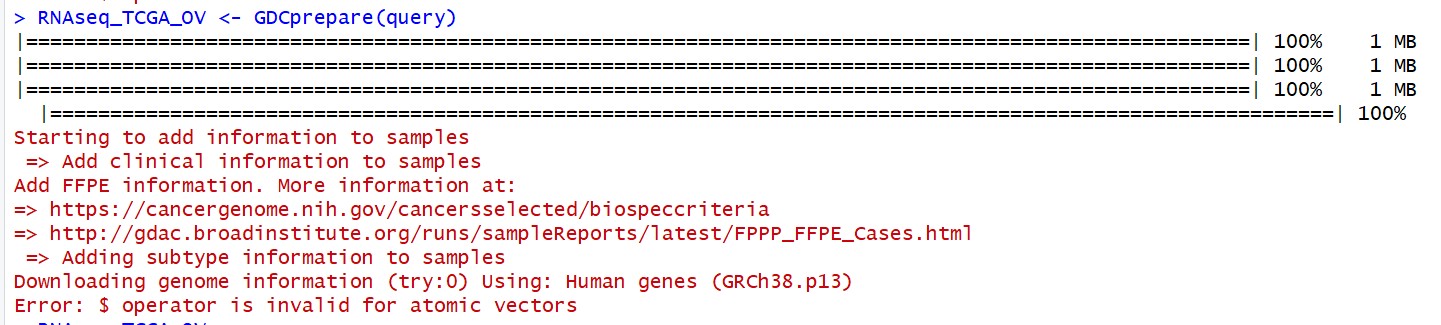
3 TCGAbiolinks使用求助 Downloading genome information (try:0) Using: Human genes (GRCh38.p13) Error: $ operator is invalid for atomic vectors
各位前辈老师们大家好!我在使用TCGAbiolinks包的GDCprepare函数的时候,出现报错,请各位给予分析或者
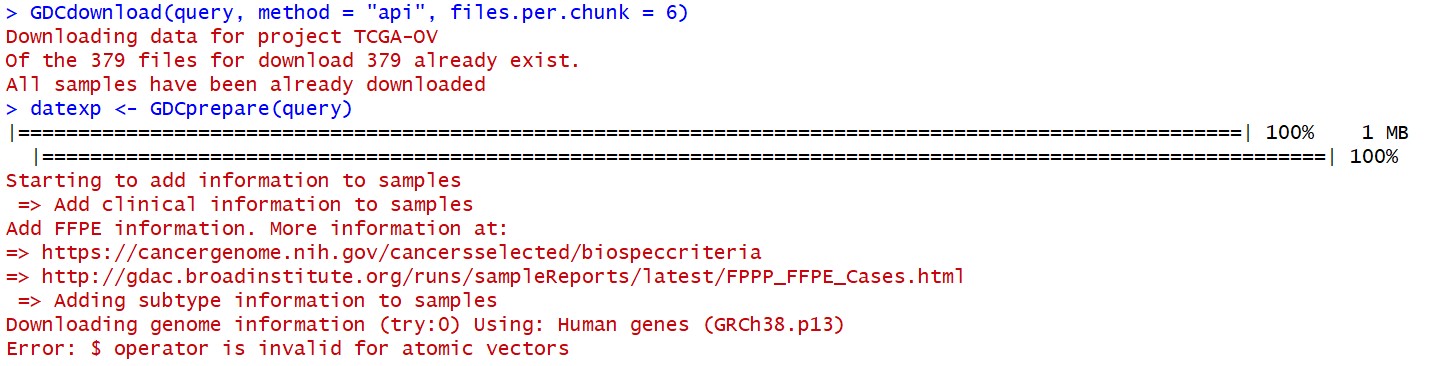
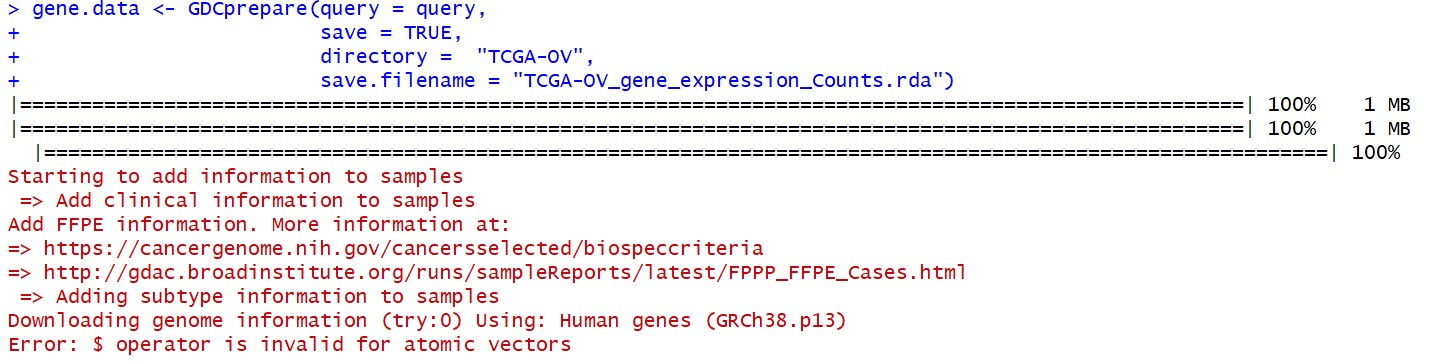
Starting to add information to samples
=> Add clinical information to samples
Add FFPE information. More information at:
=> https://cancergenome.nih.gov/cancersselected/biospeccriteria
=> http://gdac.broadinstitute.org/runs/sampleReports/latest/FPPP_FFPE_Cases.html
=> Adding subtype information to samples
Downloading genome information (try:0) Using: Human genes (GRCh38.p13)
Error: $ operator is invalid for atomic vectors
Execution halted
帮助!谢谢
问题补充:

 --------------------------------------------------------
--------------------------------------------------------
按照前辈 给的代码删除数据重新跑了一遍,结果:
请先 登录 后评论
最佳答案 2020-02-17 17:19
刚刚测试了一下,当TCGAbiolinks 为2.10.5版本的时候会报错这样的错误,应该是个bug。建议重新安装TCGAbiolinks 更新到最新版本可以解决以上问题,并且R的版本也更新到最新。
建议把下载的数据全部删除,重新下载数据后再整理,试一下。
query <- GDCquery(project = "TCGA-OV", data.category = "Transcriptome Profiling", data.type = "Gene Expression Quantification", workflow.type = "HTSeq - Counts", #不同数据解释:https://www.omicsclass.com/article/1059 legacy = FALSE) # 下载数据, 数据比较大,耐心等待 GDCdownload(query = query,directory = "TCGA-OV",files.per.chunk=10, method='api')
gene.data <- GDCprepare(query = query,
save = TRUE,
directory = "TCGA-OV",
save.filename = "TCGA-OV_gene_expression_Counts.rda")
请先 登录 后评论
如果觉得我的回答对您有用,请随意打赏。你的支持将鼓励我继续创作!

- 3 关注
- 0 收藏,6965 浏览
- 带带大学习 提出于 2020-02-08 17:44
相似问题
- 请教一下TCGA的蛋白数据怎么理解,它是有正负值的,这个正负值代表什么意思,为什么蛋白表达会有负值?我只想给它分成高表达,低表达和表达缺失组应该怎么处理呀? 0 回答
- rJava 安装包错 1 回答
- R包安装 1 回答
- R语言绘制中国地图,每个省份出现多边形 1 回答
- R 更新后报错 0 回答
- TCGA下载 docker报错 0 回答