/share/work/biosoft/TransDecoder/latest/util/gff3_file_to_proteins.pl --gff3 $species.longest_isoform.gff3 --fasta $genome --seqType prot >$species.pep.fa
这一部分的分析,提取出来的蛋白序列与官方发布的蛋白序列有出入,而且U等氨基酸字母会直接被改成"*",例如:
用脚本提取的蛋白序列其中一个序列: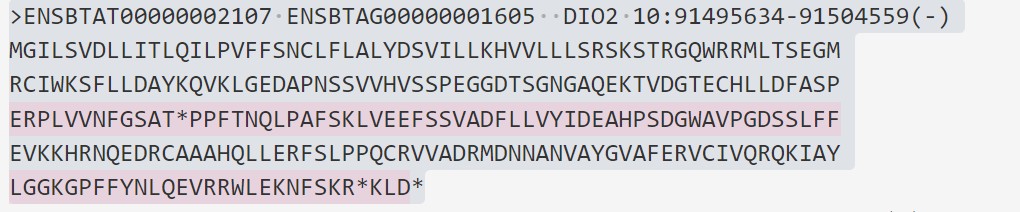
而在官网给的蛋白序列fa文件中,搜索这一条序列:
>ENSBTAP00000002107.8 pep primary_assembly:ARS-UCD1.2:10:91495432:91504569:-1 gene:ENSBTAG00000001605.8 transcript:ENSBTAT00000002107.8 gene_biotype:protein_coding transcript_biotype:protein_coding gene_symbol:DIO2 description:iodothyronine deiodinase 2 [Source:VGNC Symbol;Acc:VGNC:106710]
MGILSVDLLITLQILPVFFSNCLFLALYDSVILLKHVVLLLSRSKSTRGQWRRMLTSEGM
RCIWKSFLLDAYKQVKLGEDAPNSSVVHVSSPEGGDTSGNGAQEKTVDGTECHLLDFASP
ERPLVVNFGSATUPPFTNQLPAFSKLVEEFSSVADFLLVYIDEAHPSDGWAVPGDSSLFF
EVKKHRNQEDRCAAAHQLLERFSLPPQCRVVADRMDNNANVAYGVAFERVCIVQRQKIAY
LGGKGPFFYNLQEVRRWLEKNFSKRUKLD
这个问题怎么解决?
并且不只一个序列也不只一个基因组有这种情况

