我们的示例数据输出文件里面该文件内容如下:
建议check一下你的*_blast_results.txt是否正常,如果为空 检查 ${SPROT} 变量是否赋值
基因组重复序列分析的时候,在运行【汇总 不同软件生成的repeat库,并用RepeatMasker进行重复序列注释】时候
for lib in ModelerAll.lib MITE_LTR.lib Homology.db; do
blastx -query ${lib} -db ${SPROT} -evalue 1e-10 -num_descriptions 10 -num_threads ${threads} -out ${lib}_blast_results.txt
perl $scriptsdir/ProtExcluder1.2/ProtExcluder.pl ${lib}_blast_results.txt ${lib}
echo -e "${lib}\tbefore\t$(grep -c ">" ${lib})\tafter\t$(grep -c ">" ${lib}noProtFinal)"
done
会报错如下:
Can not open the seqfile ModelerAll.lib_blast_results.txt.fnolowm50seq
mergeunmatchedregion.pl seqfile
Illegal division by zero at /public-supool/home/thli/scripts/genome_annotation/ProtExcluder1.2/GCcontent.pl line 122.
Can not open the seqfile MITE_LTR.lib_blast_results.txt.fnolowm50seq
mergeunmatchedregion.pl seqfile
Illegal division by zero at /public-supool/home/thli/scripts/genome_annotation/ProtExcluder1.2/GCcontent.pl line 122.
Can not open the seqfile Homology.db_blast_results.txt.fnolowm50seq
mergeunmatchedregion.pl seqfile
Illegal division by zero at /public-supool/home/thli/scripts/genome_annotation/ProtExcluder1.2/GCcontent.pl line 122.
vsearch v2.18.0_linux_x86_64, 503.4GB RAM, 128 cores
A C G T N totalnoN total
00000000 00000000 00000000 00000000 00000000 00000000 00000000
AT 00000000 GC 00000000
*_blast_results.txt输出如下:
cmd>head -50 Homology.db_blast_results.txt
BLASTX 2.14.1+
Reference: Stephen F. Altschul, Thomas L. Madden, Alejandro A.
Schaffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J.
Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of
protein database search programs", Nucleic Acids Res. 25:3389-3402.
Database: uniprot_sprot_clean.fasta
565,168 sequences; 203,477,143 total letters
Query= IS1#ARTEFACT @root [S:10]
Length=768
Score E
Sequences producing significant alignments: (Bits) Value
sp|P59843|INSB_HAEDU 348 2e-122
sp|A0A385XJL4|INSB9_ECOLI 348 2e-122
sp|P0CF30|INSB8_ECOLI 348 2e-122
sp|P0CF29|INSB6_ECOLI 348 2e-122
sp|P0CF28|INSB5_ECOLI 348 2e-122
sp|P0CF25|INSB1_ECOLI 348 2e-122
sp|P0CF31|INSB_ECOLX 346 2e-121
sp|P57998|INSB4_ECOLI 338 3e-118
sp|P0CF27|INSB3_ECOLI 335 3e-117
sp|P0CF26|INSB2_ECOLI 335 3e-117
>sp|P59843|INSB_HAEDU
Length=167
Score = 348 bits (893), Expect = 2e-122, Method: Compositional matrix adjust.
Identities = 167/167 (100%), Positives = 167/167 (100%), Gaps = 0/167 (0%)
Frame = +1
Query 250 MPGNSPHYGRWPQHDFTSLKKLRPQSVTSRIQPGSDVIVCAEMDEQWGYVGAKSRQRWLF 429
MPGNSPHYGRWPQHDFTSLKKLRPQSVTSRIQPGSDVIVCAEMDEQWGYVGAKSRQRWLF
Sbjct 1 MPGNSPHYGRWPQHDFTSLKKLRPQSVTSRIQPGSDVIVCAEMDEQWGYVGAKSRQRWLF 60
Query 430 YAYDSLRKTVVAHVFGERTMATLGRLMSLLSPFDVVIWMTDGWPLYESRLKGKLHVISKR 609
YAYDSLRKTVVAHVFGERTMATLGRLMSLLSPFDVVIWMTDGWPLYESRLKGKLHVISKR
Sbjct 61 YAYDSLRKTVVAHVFGERTMATLGRLMSLLSPFDVVIWMTDGWPLYESRLKGKLHVISKR 120
Query 610 YTQRIERHNLNLRQHLARLGRKSLSFSKSVELHDKVIGHYLNIKHYQ 750
YTQRIERHNLNLRQHLARLGRKSLSFSKSVELHDKVIGHYLNIKHYQ
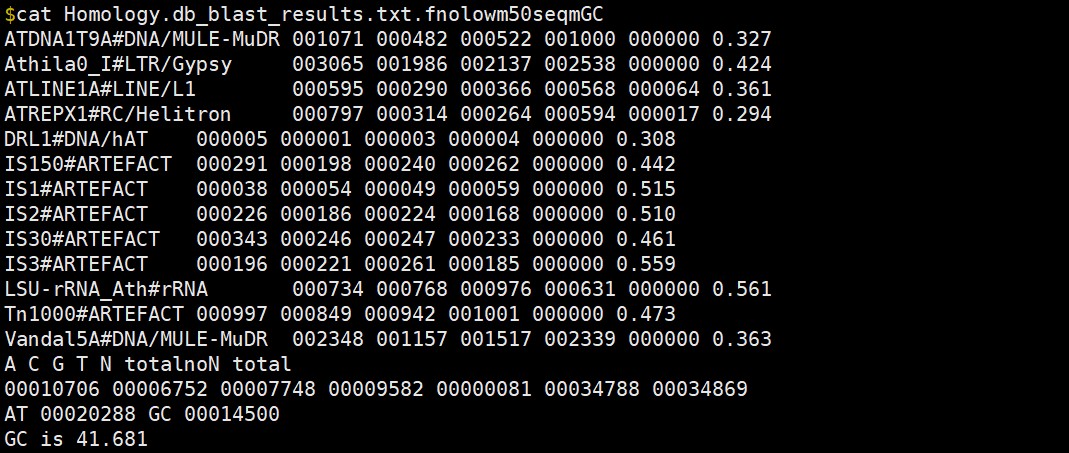
*_blast_results.txt输出正常,但是Homology.db_blast_results.txt.fnolowm50seqmGC,输出还是错误:
A C G T N totalnoN total
00000000 00000000 00000000 00000000 00000000 00000000 00000000
AT 00000000 GC 00000000