使用monocle3进行轨迹分析报错
使用单细胞转录组学课程中的代码使用monocle3进行轨迹分析报错,之前使用镜像3.0的时候没有问题,自从换成4.0后就出现了以下问题。
这是我的代码:Rscript $scripts/cell_trajectory_monocle3.r --rds ../05.Find_Markers/CD4.added.celltype.rds \
--use.seurat.umap --groupby mycelltype -o monocle3.1 --cpu 4
这是CD4.added.celltype.rds的内容:barcodeorig.identnUMInGenegrouppercent_mitopercent_hbpercent_ribolog10GenesPerUMIcellsCC.DifferenceS.ScoreG2M.ScorePhasenCount_SCTnFeature_SCTSCT_snn_res.0.8seurat_clusterspANN_0.25_0.23_3227DF.classifications_0.25_0.23_3227mycelltypeRNA_snn_res.0.3
AAACCCAAGACTAAGT-1_1pbmc_N1656339N2.28658536585366016.31097560975610.89822011771757AAACCCAAGACTAAGT-10.0133297422614858-0.0263769850155986-0.0397067272770845G13534583540.306852383285504SingletTreg4
AAACCCACACAACGAG-1_1pbmc_N132411219N7.2199938290651032.98364702252390.87903328055343AAACCCACACAACGAG-1-0.0321644920061835-0.0561094645379436-0.0239449725317602G136831219710.237418632323091SingletEffectorCD4+_T1
AAAGGATAGCCAGACA-1_1pbmc_N134491355N7.19048999710061025.86256886053930.885305514960145AAAGGATAGCCAGACA-1-0.0901341994777545-0.04724818221860860.0428860172591459G2M37451355740.27367536921677SingletTreg4
这是报错: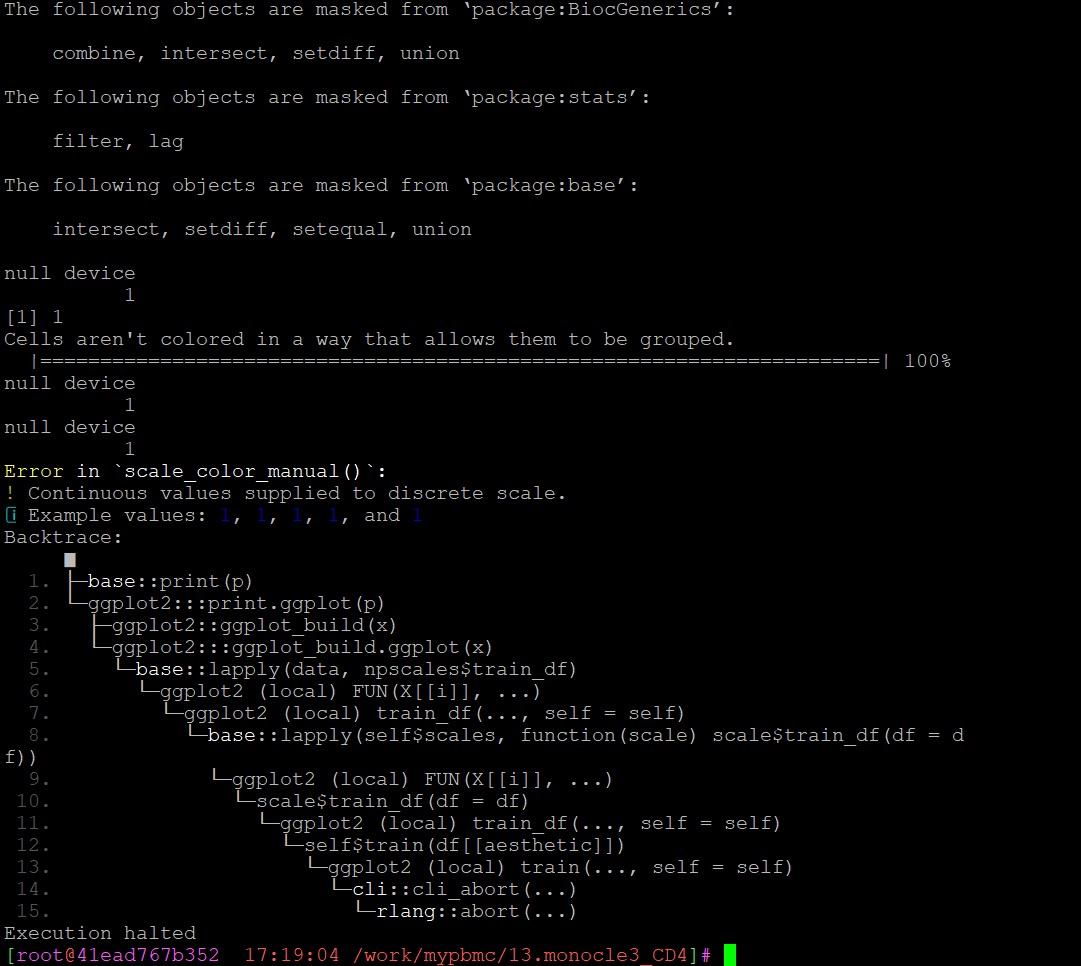
- 0 关注
- 0 收藏,51 浏览
- Zzz 提出于 3天前
相似问题
- 单细胞测序的细胞通讯分析 cellchat 1 回答
- 文件夹权限受限,无法删除 1 回答
- 单细胞测序数据两个子集合并 1 回答
- 使用monocle3进行轨迹分析报错 2 回答
- 如何合并两个单细胞测序中的两个子集? 1 回答